3GI4
 
 | | Crystal structure of protease inhibitor, KB60 in complex with wild type HIV-1 protease | | Descriptor: | 5S)-N-[(1S,2R)-3-[(1,3-Benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)propyl]-2-oxo-3-[3-(tr ifluoromethyl)phenyl]-5-oxazolidinecarboxamide, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
3GIX
 
 | |
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3GI1
 
 | | Crystal Structure of the laminin-binding protein Lbp of Streptococcus pyogenes | | Descriptor: | Laminin-binding protein of group A streptococci, ZINC ION | | Authors: | Linke, C, Caradoc-Davies, T.T, Young, P.G, Proft, T, Baker, E.N. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The laminin-binding protein Lbp from Streptococcus pyogenes is a zinc receptor
J.Bacteriol., 191, 2009
|
|
7ZX7
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4KKV
 
 | | Crystal structure of candida glabrata FMN adenylyltransferase D181A Mutant | | Descriptor: | CHLORIDE ION, Similar to uniprot|P38913 Saccharomyces cerevisiae YDL045c FAD synthetase, beta-D-glucopyranose | | Authors: | Huerta, C, Zhang, H. | | Deposit date: | 2013-05-06 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The "Super Mutant" of Yeast FMN Adenylyltransferase Enhances the Enzyme Turnover Rate by Attenuating Product Inhibition.
Biochemistry, 52, 2013
|
|
3M8D
 
 | | Crystal structure of spin-labeled BtuB V10R1 with bound calcium and cyanocobalamin | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, CYANOCOBALAMIN, ... | | Authors: | Freed, D.M, Horanyi, P.S, Wiener, M.C, Cafiso, D.S. | | Deposit date: | 2010-03-17 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational exchange in a membrane transport protein is altered in protein crystals.
Biophys.J., 99, 2010
|
|
6BAS
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
7JR7
 
 | | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8, Fab 11F4 heavy chain, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z, Zhang, H. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies
Commun Biol, 4, 2021
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4DBD
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus | | Descriptor: | Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-14 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus
To be Published
|
|
9BG0
 
 | | Tri-complex of Daraxonrasib (RMC-6236), NRAS WT, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase NRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Bieder, R, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Daraxonrasib (RMC-6236), a Potent and Orally Bioavailable RAS(ON) Multi-selective, Noncovalent Tri-complex Inhibitor for the Treatment of Patients with Multiple RAS-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
9BG3
 
 | | Tri-complex of Daraxonrasib (RMC-6236), NRAS Q61K, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, CHLORIDE ION, GTPase NRas, ... | | Authors: | Tomlinson, A.C.A, Bieder, R, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Daraxonrasib (RMC-6236), a Potent and Orally Bioavailable RAS(ON) Multi-selective, Noncovalent Tri-complex Inhibitor for the Treatment of Patients with Multiple RAS-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
6IBY
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 6 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpyrrolidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7K2D
 
 | | Kelch domain of human KEAP1 bound to Nrf2 linear peptide, Ac-GDEETGE-NH2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 linear peptide, Ace-GDEETGE-NH2 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
4K9Z
 
 | | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P6222 | | Descriptor: | CHLORIDE ION, Putative thiol-disulfide oxidoreductase | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R.D, Bonanno, J.B, Armstrong, R.N, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P6222
To be Published
|
|
4DWO
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-26 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II
To be Published
|
|
4KAN
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)acetic acid, ADENOSINE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid
To be Published
|
|
6IL7
 
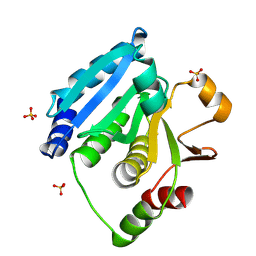 | | Structure of Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F (AhpF) C503A mutant | | Descriptor: | SULFATE ION, Thioredoxin reductase/glutathione-related protein | | Authors: | Toh, Y.K, Shin, J, Balakrishna, A.M, Eisenhaber, F, Eisenhaber, B, Gruber, G. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of the additional cysteine 503 of vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F (AhpF) and the mechanism of AhpF and subunit C assembling.
Free Radic. Biol. Med., 138, 2019
|
|
6BJ9
 
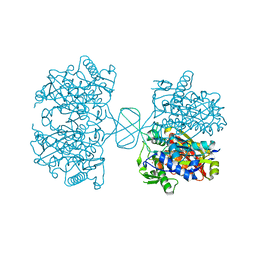 | |
3ME7
 
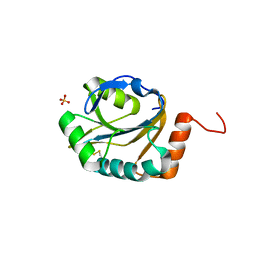 | | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
2H36
 
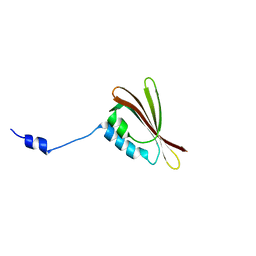 | |
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4E02
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1547 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
3MF8
 
 | | Crystal Structure of Native cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
