5NBW
 
 | |
5EW9
 
 | | Crystal Structure of Aurora A Kinase Domain Bound to MK-5108 | | Descriptor: | 4-(3-chloranyl-2-fluoranyl-phenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid, Aurora kinase A | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | A Cell Biologist's Field Guide to Aurora Kinase Inhibitors.
Front Oncol, 5, 2015
|
|
2PVK
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-CHLOROBENZYLAMINO)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3AOC
 
 | |
1Z19
 
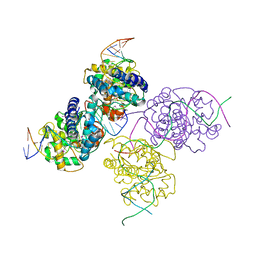 | | Crystal structure of a lambda integrase(75-356) dimer bound to a COC' core site | | Descriptor: | 33-MER, 5'-D(*CP*TP*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*AP*TP*AP*CP*TP*AP*AP*GP*TP*TP*GP*GP*CP*AP*TP*TP*A)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
2VQQ
 
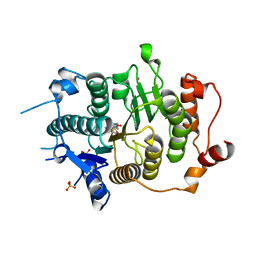 | | Structure of HDAC4 catalytic domain (a double cysteine-to-alanine mutant) bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQO
 
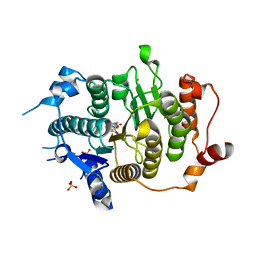 | | Structure of HDAC4 catalytic domain with a gain-of-function muation bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
5SFH
 
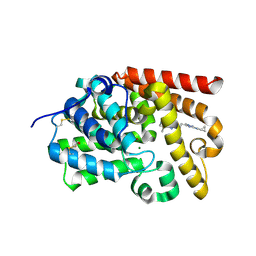 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N(C1CC1)(C)c2nn(c(n2)CCc4nn3c(cnc(c3n4)C)C)C, micromolar IC50=0.042255 | | Descriptor: | MAGNESIUM ION, N-cyclopropyl-5-{2-[(4S)-5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl]ethyl}-N,1-dimethyl-1H-1,2,4-triazol-3-amine, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFS
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2c(C)c1nc(nn1c(c2)C)CCc3nc(nn3C)N4CC[C@@H](C4)C, micromolar IC50=0.012855 | | Descriptor: | (4S)-5,8-dimethyl-2-(2-{1-methyl-3-[(3S)-3-methylpyrrolidin-1-yl]-1H-1,2,4-triazol-5-yl}ethyl)[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFY
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2c(c1nc(nn1c(c2)C)C=Cc3nc(nn3C)c4c[nH]nc4)C, micromolar IC50=0.22176 | | Descriptor: | (4S)-5,8-dimethyl-2-{(E)-2-[1-methyl-3-(1H-pyrazol-4-yl)-1H-1,2,4-triazol-5-yl]ethenyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SER
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(cnn1C)C(=O)N(C)CCF)C(Nc3cc2nc(nn2cc3)c4ccccc4)=O, micromolar IC50=0.0009363 | | Descriptor: | MAGNESIUM ION, N~4~-(2-fluoroethyl)-N~4~,1-dimethyl-N~5~-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SE3
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(cc1nc(nn1cc2)c3ccccc3)NC(c4ccnc(c4)Cl)=O, micromolar IC50=0.045025 | | Descriptor: | 2-chloro-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]pyridine-4-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFD
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1(nc(nc1CCc2nn3c(n2)c(ncc3C)C)N4CCCCC4)C, micromolar IC50=0.03165 | | Descriptor: | (4S)-5,8-dimethyl-2-{2-[1-methyl-3-(piperidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFI
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1(nc(nc1CCc2nn3c(n2)c(ncc3C)C)N4[C@H](CCC4)C)C, micromolar IC50=0.028433 | | Descriptor: | (4S)-5,8-dimethyl-2-(2-{1-methyl-3-[(2S)-2-methylpyrrolidin-1-yl]-1H-1,2,4-triazol-5-yl}ethyl)[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SF7
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c2c(ccc1OC)C(=O)N(c3c(nc(n23)c4c(Cl)cccc4)C)C, micromolar IC50=0.0075033 | | Descriptor: | (10S)-1-(2-chlorophenyl)-8-methoxy-3,4-dimethylimidazo[1,5-a]quinazolin-5(4H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SE7
 
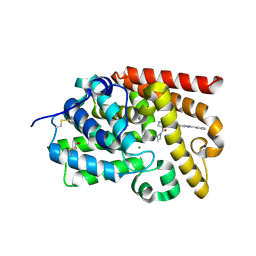 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(=O)N1CCC1)cn2)C(Nc4cc3nc(nn3cc4)c5ccccc5)=O)C, micromolar IC50=0.00054035 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFP
 
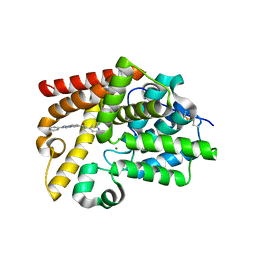 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c3(cn1c(cc(n1)c2ccccc2)cc3)C(=O)Nc4ccnc(c4)Cl, micromolar IC50=0.036749 | | Descriptor: | (8S)-N-(2-chloropyridin-4-yl)-2-phenylpyrazolo[1,5-a]pyridine-6-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6AOS
 
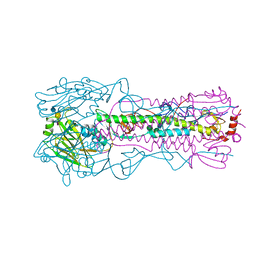 | |
6AOQ
 
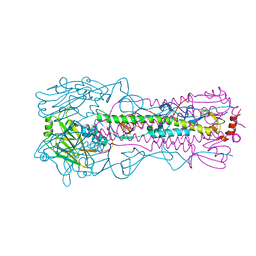 | |
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | Descriptor: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 32, 2024
|
|
6RHZ
 
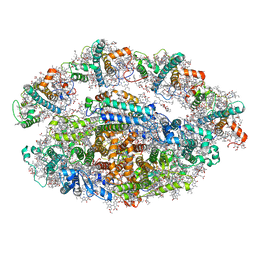 | | Structure of a minimal photosystem I from a green alga | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Perez Boerema, A, Klaiman, D, Caspy, I, Netzer-El, S.Y, Amunts, A, Nelson, N. | | Deposit date: | 2019-04-23 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
1AXO
 
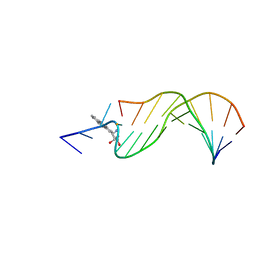 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
3GID
 
 | |
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
6AOR
 
 | |
