2DVA
 
 | | Crystal structure of peanut lectin GAL-BETA-1,3-GALNAC-ALPHA-O-ME (Methyl-T-antigen) complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3R1S
 
 | | CDK2 in complex with inhibitor KVR-1-127 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(6-chloropyridin-3-yl)methyl]amino}-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
4K9H
 
 | | Bace-1 inhibitor complex | | Descriptor: | 1-cyclopentyl-N-[(2S,3R)-3-hydroxy-1-phenyl-4-{[3-(trifluoromethyl)benzyl]amino}butan-2-yl]-6-oxo-5-(2-oxopyrrolidin-1-yl)-1,6-dihydropyridine-3-carboxamide, Beta-secretase 1 | | Authors: | Jordan, S.R. | | Deposit date: | 2013-04-19 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2EWB
 
 | | The crystal structure of Bovine Lens Leucine Aminopeptidase in complex with zofenoprilat | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Alterio, V, Pedone, C, De Simone, G. | | Deposit date: | 2005-11-02 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal Ion Substitution in the Catalytic Site Greatly Affects the Binding of Sulfhydryl-Containing Compounds to Leucyl Aminopeptidase.
Biochemistry, 45, 2006
|
|
2P8Q
 
 | |
2PN1
 
 | |
2PQX
 
 | | E. coli RNase 1 (in vivo folded) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Ribonuclease I | | Authors: | Messens, J, Loris, R, Wyns, L. | | Deposit date: | 2007-05-03 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of E. coli ribonuclease 1
To be Published
|
|
2Q1B
 
 | | Carbonic Anhydrase II in Complex with Saccharin | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Klebe, G, Heine, A, Koehler, K. | | Deposit date: | 2007-05-24 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Saccharin Inhibits Carbonic Anhydrases: Possible Explanation for its Unpleasant Metallic Aftertaste.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2PMA
 
 | | Structural Genomics, the crystal structure of a protein Lpg0085 with unknown function (DUF785) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | ACETATE ION, FORMIC ACID, Uncharacterized protein | | Authors: | Tan, K, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The crystal structure of a protein Lpg0085 with unknown function (DUF785) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
2CZ3
 
 | | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-2 crystal) | | Descriptor: | Maleylacetoacetate isomerase | | Authors: | Mizohata, E, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-2 crystal)
To be Published
|
|
1A47
 
 | | CGTASE FROM THERMOANAEROBACTERIUM THERMOSULFURIGENES EM1 IN COMPLEX WITH A MALTOHEXAOSE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1998-02-11 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Engineering of cyclodextrin product specificity and pH optima of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1.
J.Biol.Chem., 273, 1998
|
|
2K4U
 
 | | Solution structure of the SCORPION TOXIN ADWX-1 | | Descriptor: | Potassium channel toxin alpha-KTx 3.6 | | Authors: | Yin, S.J, Jiang, L, Yi, H, Han, S, Yang, D.W, Liu, M.L, Liu, H, Cao, Z.J, Wu, Y.L, Li, W.X. | | Deposit date: | 2008-06-18 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Different Residues in Channel Turret Determining the Selectivity of ADWX-1 Inhibitor Peptide between Kv1.1 and Kv1.3 Channels
J.Proteome Res., 7, 2008
|
|
1E5N
 
 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
3GLY
 
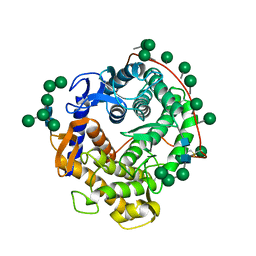 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-06-03 | | Release date: | 1994-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
2BKZ
 
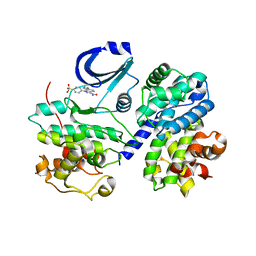 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-404611 | | Descriptor: | 1-[4-(AMINOSULFONYL)PHENYL]-1,6-DIHYDROPYRAZOLO[3,4-E]INDAZOLE-3-CARBOXAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | DAlessio, R, Bargiottia, A, Metz, S, Brasca, M.G, Cameron, A, Ermoli, A, Marsiglio, A, Polucci, P, Roletto, F, Tibolla, M, Vazquez, M.L, Vulpetti, A, Pevarello, P. | | Deposit date: | 2005-02-23 | | Release date: | 2006-03-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Benzodipyrazoles: A New Class of Potent Cdk2 Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BWA
 
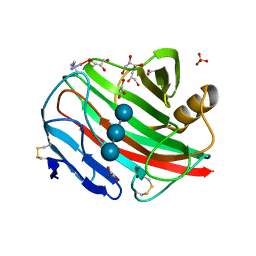 | |
2KTP
 
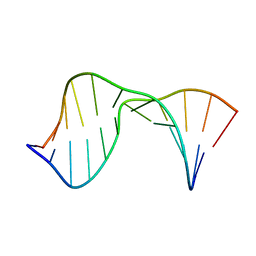 | | Structure of the 1,N2-ethenodeoxyguanosine lesion opposite a one-base deletion in duplex DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*(GNE)P*GP*AP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*TP*CP*AP*TP*GP*CP*G)-3') | | Authors: | Shanmugam, G, Kozekov, I.D, Guengerich, P.F, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the 1,N(2)-etheno-2'-deoxyguanosine lesion in the 3'-G(epsilon dG)T-5' sequence opposite a one-base deletion.
Biochemistry, 49, 2010
|
|
2KTC
 
 | | Solution Structure of a Novel hKv1.1 inhibiting scorpion toxin from Mesibuthus tamulus | | Descriptor: | Potassium channel toxin alpha-KTx 9.4 | | Authors: | Kumar, G.S, Upadhyay, S, Mathew, M.K, Sarma, S.P. | | Deposit date: | 2010-01-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BTK-2, a novel hK(v)1.1 inhibiting scorpion toxin, from the eastern Indian scorpion Mesobuthus tamulus.
Biochim.Biophys.Acta, 1814, 2011
|
|
2C9D
 
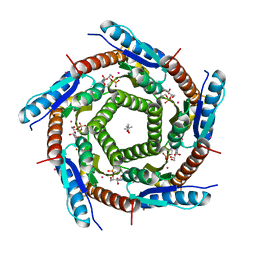 | | Lumazine Synthase from Mycobacterium tuberculosis Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL)HEXANE 1-PHOSPHATE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL ) HEXANE 1-PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
3SW2
 
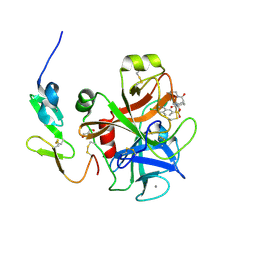 | | X-ray crystal structure of human FXA in complex with 6-chloro-N-((3S)-2-oxo-1-(2-oxo-2-((5S)-8-oxo-5,6-dihydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-3(2H,4H,8H)-yl)ethyl)piperidin-3-yl)naphthalene-2-sulfonamide | | Descriptor: | 6-chloro-N-((3S)-2-oxo-1-(2-oxo-2-((5S)-8-oxo-5,6-dihydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-3(2H,4H,8H)-yl)ethyl)piperidin-3-yl)naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-13 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Arylsulfonamidopiperidone derivatives as a novel class of factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2KNE
 
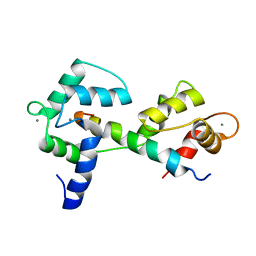 | | Calmodulin wraps around its binding domain in the plasma membrane CA2+ pump anchored by a novel 18-1 motif | | Descriptor: | ATPase, Ca++ transporting, plasma membrane 4, ... | | Authors: | Juranic, N, Atanasova, E, Filoteo, A.G, Macura, S, Prendergast, F.G, Penniston, J.T, Strehler, E.E. | | Deposit date: | 2009-08-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif.
J.Biol.Chem., 285, 2010
|
|
3OLI
 
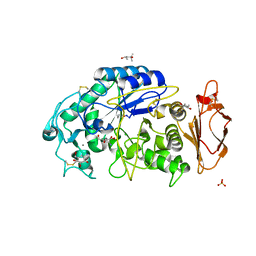 | | Structures of human pancreatic alpha-amylase in complex with acarviostatin IV03 | | Descriptor: | (1S,2S,3R,6R)-6-amino-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Qin, X, Ren, L. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes.
J.Struct.Biol., 174, 2011
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
1H23
 
 | | Structure of acetylcholinesterase (E.C. 3.1.1.7) complexed with (S,S)-(-)-bis(12)-hupyridone at 2.15A resolution | | Descriptor: | (S,S)-(-)-N,N'-DI-5'-[5',6',7',8'-TETRAHYDRO- 2'(1'H)-QUINOLYNYL]-1,12-DIAMINODODECANE DIHYDROCHLORIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.F, Pang, Y.P, Silman, I, Sussman, J.L. | | Deposit date: | 2002-07-30 | | Release date: | 2002-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Acetylcholinesterase Complexed with Bivalent Ligands Related to Huperzine A: Experimental Evidence for Species-Dependent Protein-Ligand Complementarity
J.Am.Chem.Soc., 125, 2003
|
|
2LIR
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in oxidized states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
