7B0N
 
 | |
6G8G
 
 | | Flavonoid-responsive Regulator FrrA in complex with Genistein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GENISTEIN, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
6MTK
 
 | |
2AQ9
 
 | | Structure of E. coli LpxA with a bound peptide that is competitive with acyl-ACP | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Williams, A.H, Immormino, R.M, Gewirth, D.T, Raetz, C.R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3LC0
 
 | |
2AT3
 
 | |
5YJS
 
 | | Structure of vicilin from Capsicum annuum | | Descriptor: | 2-HYDROXYBENZOIC ACID, CHLORIDE ION, COPPER (I) ION, ... | | Authors: | Shikhi, M, Nair, D.T, Salunke, D.M. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-guided identification of function: role ofCapsicum annuumvicilin during oxidative stress.
Biochem. J., 475, 2018
|
|
7X39
 
 | | Structure of CIZ1 bound ERH | | Descriptor: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
6GHS
 
 | | Modification dependent TagI restriction endonuclease | | Descriptor: | SODIUM ION, TagI restriction endonuclease, ZINC ION | | Authors: | Kisiala, M, Copelas, A, Czapinska, H, Xu, S, Bochtler, M. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of the modification-dependent SRA-HNH endonuclease TagI.
Nucleic Acids Res., 46, 2018
|
|
4R26
 
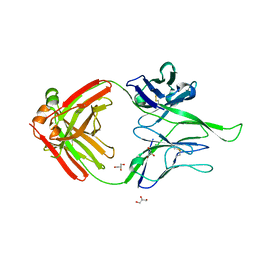 | | Crystal structure of human Fab PGT124, a broadly neutralizing and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGR124-Light Chain, PGT124-Heavy Chain | | Authors: | Garces, F, Kong, L, Wilson, I.A. | | Deposit date: | 2014-08-08 | | Release date: | 2014-10-08 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.4969 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
4R2G
 
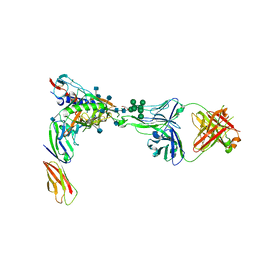 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
1AR1
 
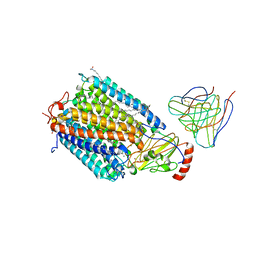 | | Structure at 2.7 Angstrom Resolution of the Paracoccus Denitrificans two-subunit Cytochrome C Oxidase Complexed with an Antibody Fv Fragment | | Descriptor: | ANTIBODY FV FRAGMENT, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Ostermeier, C, Harrenga, A, Ermler, U, Michel, H. | | Deposit date: | 1997-08-08 | | Release date: | 1998-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5DDP
 
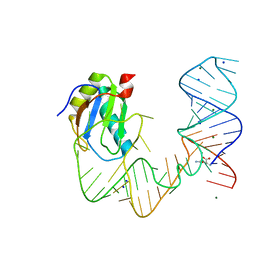 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
4WZ8
 
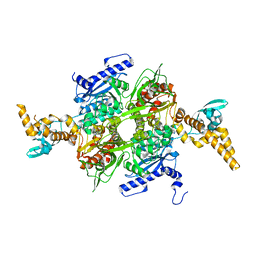 | |
4WYO
 
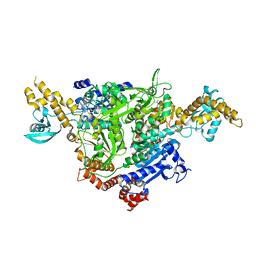 | |
2IPG
 
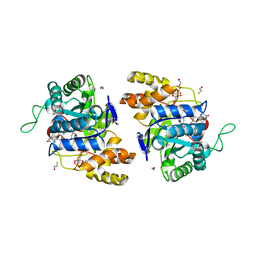 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase mutant K31A in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, 1,2-ETHANEDIOL, 3(17)alpha-hydroxysteroid dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Lemieux, M, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
8FJV
 
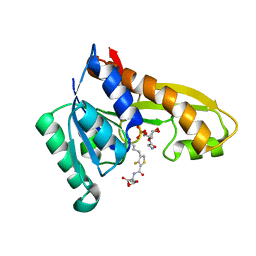 | | Human GAR transformylase in complex with GAR substrate and AGF362 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-3-fluorothiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
8FJX
 
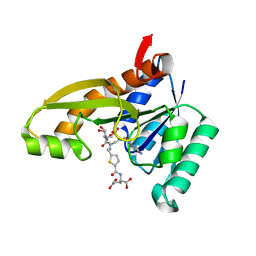 | | Human GAR transformylase in complex with GAR substrate and AGF320 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[5-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)pentyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
8FJY
 
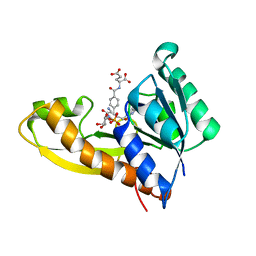 | | Human GAR transformylase in complex with GAR substrate and AGF291 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[3-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)propyl]benzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
8FJW
 
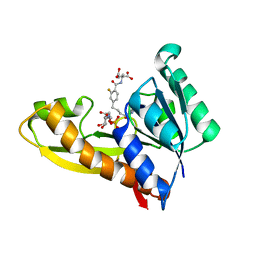 | | Human GAR transformylase in complex with GAR substrate and AGF347 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
6TD2
 
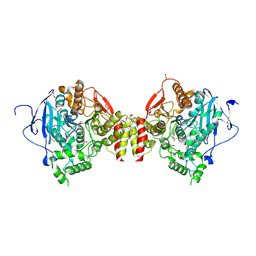 | |
2IPF
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, (3(17)alpha-hydroxysteroid dehydrogenase), 1,2-ETHANEDIOL, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
4JAZ
 
 | | Crystal structure of the complex between PPARgamma LBD and trans-resveratrol | | Descriptor: | Peroxisome proliferator-activated receptor gamma, RESVERATROL | | Authors: | Pochetti, G, Capelli, D, Montanari, R, Calleri, E, Moaddel, R, Temporini, C. | | Deposit date: | 2013-02-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Resveratrol and Its Metabolites Bind to PPARs.
Chembiochem, 15, 2014
|
|
6G87
 
 | | Flavonoid-responsive Regulator FrrA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-24 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
7RQZ
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 48 degrees Celsius, in an open state, class alpha | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
