5ZCA
 
 | | Crystal structure of lambda repressor (1-20) fused with maltose-binding protein | | Descriptor: | CITRIC ACID, Repressor protein cI,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-02-16 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Co-translational folding of alpha-helical proteins: structural studies of intermediate-length variants of the lambda repressor
Febs Open Bio, 8, 2018
|
|
3E2R
 
 | |
4ND5
 
 | |
4ND2
 
 | |
4NXJ
 
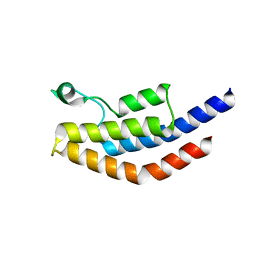 | | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum | | Descriptor: | Bromodomain protein | | Authors: | Wernimont, A.K, Loppnau, P, Knapp, S, Fonseca, M, Brennan, P.E, Dong, A, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum
TO BE PUBLISHED
|
|
4ND1
 
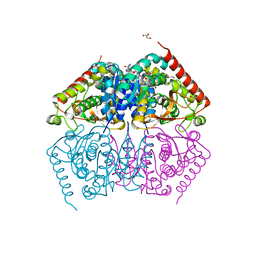 | |
5ZB6
 
 | |
4NUF
 
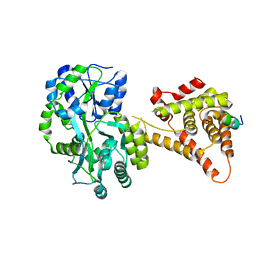 | | Crystal Structure of SHP/EID1 | | Descriptor: | EID1 peptide, Maltose ABC transporter periplasmic protein, Nuclear receptor subfamily 0 group B member 2 chimeric construct, ... | | Authors: | Zhi, X, Zhou, X.E, He, Y, Zechner, C, Suino-Powell, K.M, Kliewer, S.A, Melcher, K, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into gene repression by the orphan nuclear receptor SHP.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CLD
 
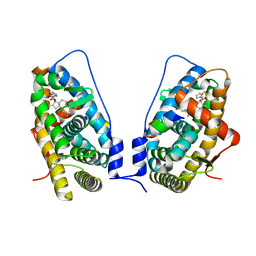 | | Ligand binding domain of the glucocorticoid receptor complexed with fluticazone furoate | | Descriptor: | (6alpha,11alpha,14beta,16alpha,17alpha)-6,9-difluoro-17-{[(fluoromethyl)sulfanyl]carbonyl}-11-hydroxy-16-methyl-3-oxoan drosta-1,4-dien-17-yl furan-2-carboxylate, Glucocorticoid receptor, Tif2 coactivator motif | | Authors: | Shewchuk, L.M, McLay, I, Stewart, E, Biggadike, K.B, Hassell, A.M, Bledsoe, R.K. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | X-ray crystal structure of the novel enhanced-affinity glucocorticoid agonist fluticasone furoate in the glucocorticoid receptor-ligand binding domain.
J.Med.Chem., 51, 2008
|
|
2IWG
 
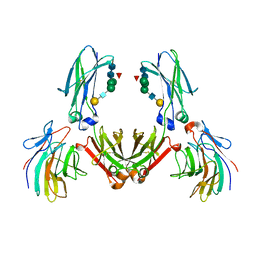 | | COMPLEX BETWEEN THE PRYSPRY DOMAIN OF TRIM21 AND IGG FC | | Descriptor: | 52 KDA RO PROTEIN, IG GAMMA-1 CHAIN C, alpha-L-fucopyranose, ... | | Authors: | James, L.C, Keeble, A.H, Rhodes, D.A, Trowsdale, J. | | Deposit date: | 2006-06-30 | | Release date: | 2007-03-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Pryspry-Mediated Tripartite Motif (Trim) Protein Function.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2Y5L
 
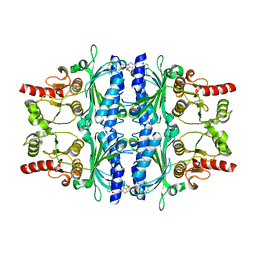 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4ND3
 
 | |
4NYA
 
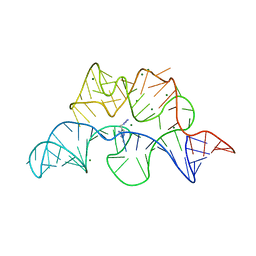 | | Crystal structure of the E. coli thiM riboswitch in complex with 5-(azidomethyl)-2-methylpyrimidin-4-amine | | Descriptor: | 5-(azidomethyl)-2-methylpyrimidin-4-amine, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
2J0S
 
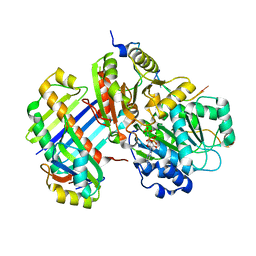 | | The crystal structure of the Exon Junction Complex at 2.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP *UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
4JL9
 
 | | Crystal structure of mouse TBK1 bound to BX795 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P, Shu, C. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.0999 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
4JNN
 
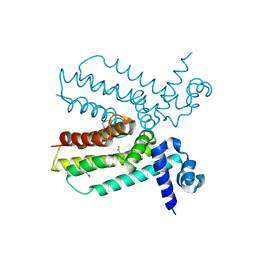 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine | | Descriptor: | BENZAMIDINE, BETA-MERCAPTOETHANOL, Transcriptional regulator | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-15 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine
To be Published
|
|
5ZVQ
 
 | |
4DNC
 
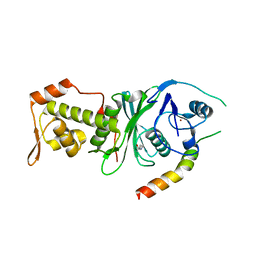 | | Crystal structure of human MOF in complex with MSL1 | | Descriptor: | Histone acetyltransferase KAT8, Male-specific lethal 1 homolog, ZINC ION | | Authors: | Huang, J, Lei, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the regulation of MOF in the male-specific lethal complex and the non-specific lethal complex.
Cell Res., 22, 2012
|
|
2FHY
 
 | |
2VUX
 
 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
4KNW
 
 | |
4ME9
 
 | |
2J7J
 
 | |
4LY7
 
 | | Ancestral RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Hart, K.M, Harms, M.J, Schmidt, B.H, Elya, C, Thornton, J.W, Marqusee, S. | | Deposit date: | 2013-07-30 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Thermodynamic system drift in protein evolution.
Plos Biol., 12, 2014
|
|
3D24
 
 | | Crystal structure of ligand-binding domain of estrogen-related receptor alpha (ERRalpha) in complex with the peroxisome proliferators-activated receptor coactivator-1alpha box3 peptide (PGC-1alpha) | | Descriptor: | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR1 | | Authors: | Moras, D, Greschik, H, Flaig, R, Sato, Y, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Communication between the ERR{alpha} Homodimer Interface and the PGC-1{alpha} Binding Surface via the Helix 8-9 Loop.
J.Biol.Chem., 283, 2008
|
|
