8TOQ
 
 | | ACE2-peptide 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOO
 
 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TOM
 
 | |
8TOL
 
 | |
8TOK
 
 | |
8TOI
 
 | |
8TOF
 
 | | Rpd3S bound to an H3K36Cme3 modified nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (176-MER), Histone H2A, ... | | Authors: | Markert, J.W, Vos, S.M, Farnung, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex.
Nat Commun, 14, 2023
|
|
8TOE
 
 | |
8TOC
 
 | | Acinetobacter phage AP205 | | Descriptor: | Coat protein, Maturation protein, RNA (4269-MER) | | Authors: | Meng, R, Xing, Z, Chang, J, Zhang, J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TOB
 
 | | Acinetobacter GP16 Type IV pilus | | Descriptor: | Fimbrial protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TOA
 
 | | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H7.HK2 Neutralizing Antibody Heavy Chain, H7.HK2 Neutralizing Antibody Light Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Wu, X, Shapiro, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK1
To Be Published
|
|
8TO8
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO6
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO5
 
 | | Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TO2
 
 | | Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TO1
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO0
 
 | | 48-nm repeating structure of doublets from mouse sperm flagella | | Descriptor: | Cilia- and flagella- associated protein 210, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Chen, Z, Shiozak, M, Hass, K.M, Skinner, W, Zhao, S, Guo, C, Polacco, B.J, Yu, Z, Krogan, N.J, Kaake, R.M, Vale, R.D, Agard, D.A. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking.
Cell, 186, 2023
|
|
8TNY
 
 | |
8TNX
 
 | |
8TNW
 
 | |
8TNV
 
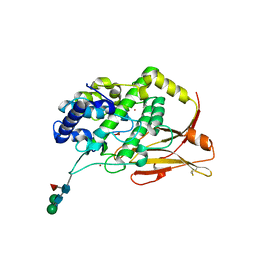 | | Hemocyanin Functional Unit CCHB-g of Concholepas concholepas | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Munoz, S, Vallejos-Baccelliere, G, Manubens, A, Salazar, M, Nascimento, A.F.Z, Ambrosio, A.L.B, Becker, M.I, Guixe, V, Castro-Fernandez, V. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a functional unit from an immunogenic mollusk hemocyanin.
Structure, 32, 2024
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNS
 
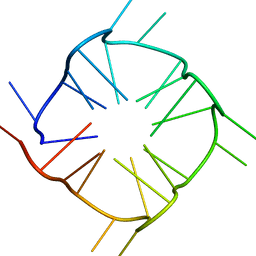 | |
8TNR
 
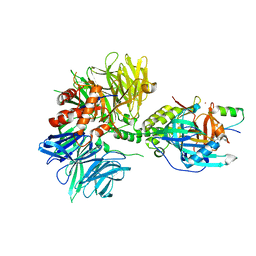 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNQ
 
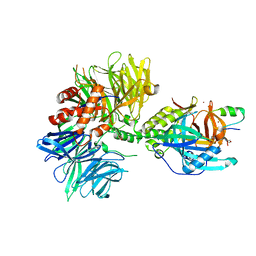 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
