1FJG
 
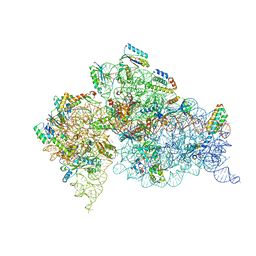 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
1Y60
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 with bound 5,10-methylene tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Formaldehyde-activating enzyme fae | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
2Y0M
 
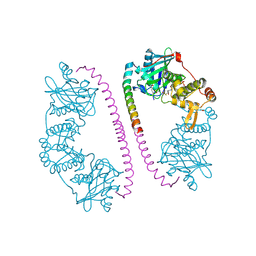 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MOF | | Descriptor: | ACETYL COENZYME *A, MALE-SPECIFIC LETHAL 1 HOMOLOG, PROBABLE HISTONE ACETYLTRANSFERASE MYST1, ... | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2Y0N
 
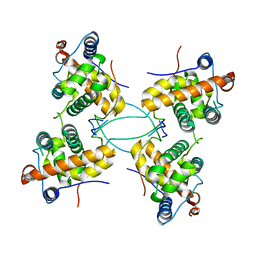 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MSL3 | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 3 HOMOLOG | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-04 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3EQG
 
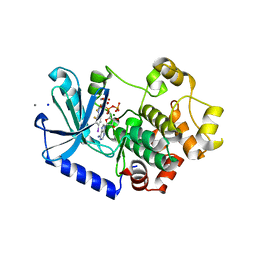 | |
2OD1
 
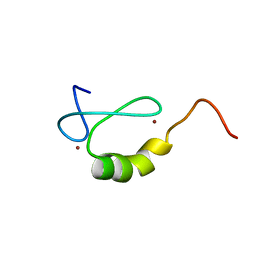 | | Solution structure of the MYND domain from human AML1-ETO | | Descriptor: | Protein CBFA2T1, ZINC ION | | Authors: | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
8SON
 
 | |
1FSI
 
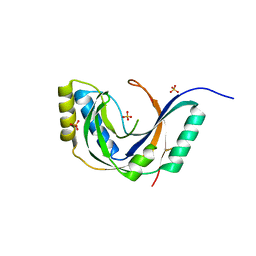 | | CRYSTAL STRUCTURE OF CYCLIC NUCLEOTIDE PHOSPHODIESTERASE OF APPR>P FROM ARABIDOPSIS THALIANA | | Descriptor: | CYCLIC PHOSPHODIESTERASE, SULFATE ION | | Authors: | Hofmann, A, Zdanov, A, Genschik, P, Filipowicz, W, Ruvinov, S, Wlodawer, A. | | Deposit date: | 2000-09-10 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction.
EMBO J., 19, 2000
|
|
2CKW
 
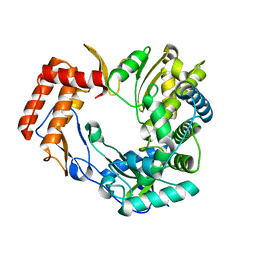 | | The 2.3 A resolution structure of the Sapporo virus RNA dependant RNA polymerase. | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Fullerton, S.W.B, Tucker, P.A, Rohayem, J, Coutard, B, Gebhardt, J, Gorbalenya, A, Canard, B. | | Deposit date: | 2006-04-24 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Sapovirus RNA-Dependent RNA Polymerase.
J.Virol., 81, 2007
|
|
1Y73
 
 | | HIV-1 Dis(Mal) Duplex Pt-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, PLATINUM (IV) ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
3UFE
 
 | | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grosse, C, Himmel, S, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2011-11-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution
To be Published
|
|
2CR7
 
 | |
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
2NYI
 
 | | Crystal Structure of an Unknown Protein from Galdieria sulphuraria | | Descriptor: | unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tandem ACT domain-containing protein ACTP from Galdieria sulphuraria.
Proteins, 80, 2012
|
|
1Z3L
 
 | | X-Ray Crystal Structure of a Mutant Ribonuclease S (F8Anb) | | Descriptor: | Ribonuclease pancreatic, S-Peptide, S-Protein, ... | | Authors: | Das, M, Vasudeva Rao, B, Ghosh, S, Varadarajan, R. | | Deposit date: | 2005-03-14 | | Release date: | 2005-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Attempts to delineate the relative contributions of changes in hydrophobicity and packing to changes in stability of ribonuclease S mutants.
Biochemistry, 44, 2005
|
|
2ZU6
 
 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3UIG
 
 | |
3UIP
 
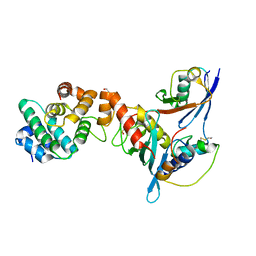 | | Complex between human RanGAP1-SUMO1, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
1Z9D
 
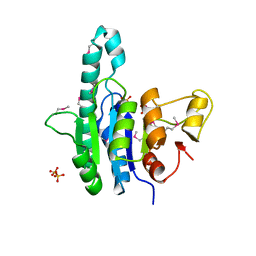 | | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes | | Descriptor: | SULFATE ION, uridylate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes
To be Published
|
|
3UR0
 
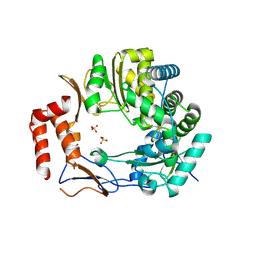 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
2O9B
 
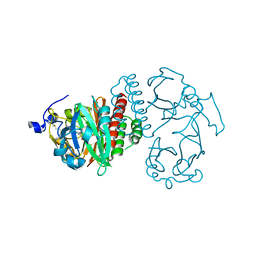 | | Crystal Structure of Bacteriophytochrome chromophore binding domain | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Vierstra, R.D, Forest, K.T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | High resolution structure of deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution.
J.Biol.Chem., 282, 2007
|
|
7JVU
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) I45T mutation complexed with SAHA | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5004766 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
1ZAI
 
 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from rabbit muscle | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZCH
 
 | | Structure of the hypothetical oxidoreductase YcnD from Bacillus subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morokutti, A, Lyskowski, A, Sollner, S, Pointner, E, Fitzpatrick, T.B, Kratky, C, Gruber, K, Macheroux, P. | | Deposit date: | 2005-04-12 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Function of YcnD from Bacillus subtilis, a Flavin-Containing Oxidoreductase(,).
Biochemistry, 44, 2005
|
|
7NC4
 
 | |
