8TQP
 
 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-{(1R)-2-(3,5-difluorophenyl)-1-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}acetamide, Gag polyprotein | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
8TQO
 
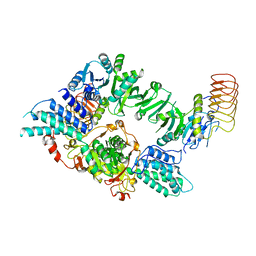 | | Eukaryotic translation initiation factor 2B tetramer | | Descriptor: | Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, Translation initiation factor eIF-2B subunit epsilon, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
8TQM
 
 | |
8TQK
 
 | |
8TQI
 
 | |
8TQG
 
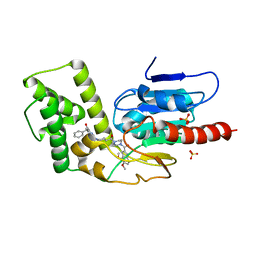 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20419 | | Descriptor: | N-benzyl-2-{4-[4-(4,5-dimethoxy-1H-indole-2-carbonyl)piperazine-1-carbonyl]piperidin-1-yl}-6-methylpyrimidine-4-carboxamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TQE
 
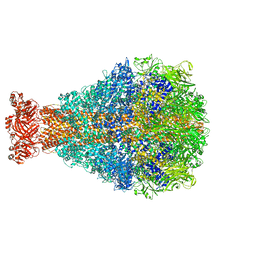 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Binshtein, E.M, Aller, S.G. | | Deposit date: | 2023-08-07 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
8TQD
 
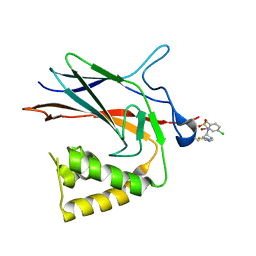 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
8TQA
 
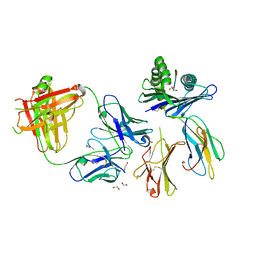 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ9
 
 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | Descriptor: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ8
 
 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ0
 
 | |
8TPY
 
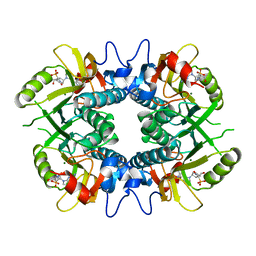 | |
8TPV
 
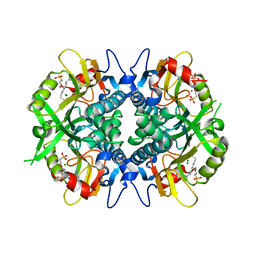 | |
8TPT
 
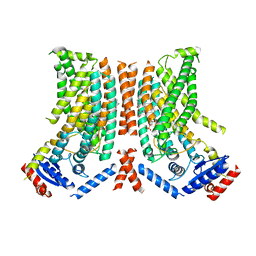 | |
8TPS
 
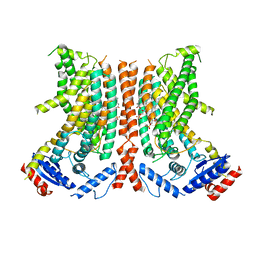 | |
8TPR
 
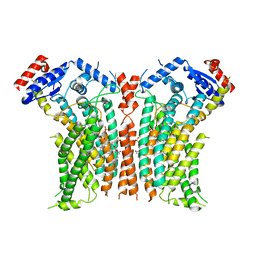 | |
8TPQ
 
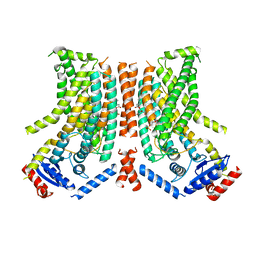 | |
8TPP
 
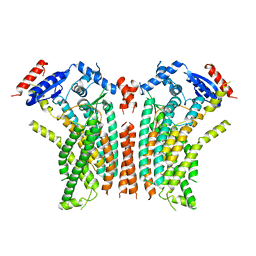 | |
8TPO
 
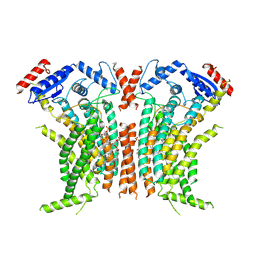 | |
8TPN
 
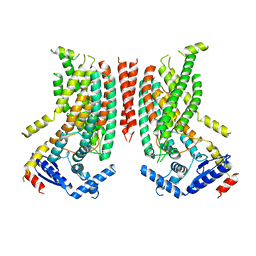 | |
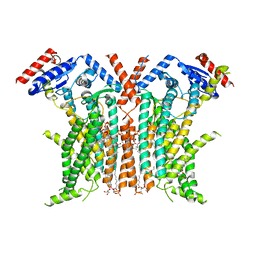
8TPM
 
 | |
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TPK
 
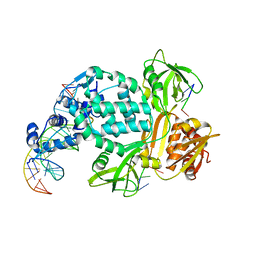 | | P6522 crystal form of C. crescentus DriD-ssDNA-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
