6AJJ
 
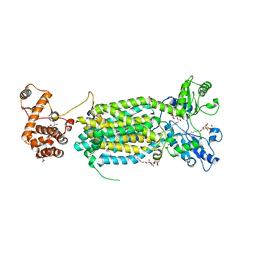 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
4QI5
 
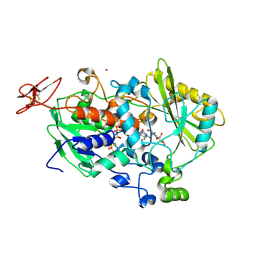 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase with bound cellobionolactam, MtDH | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4LS4
 
 | | Crystal structure of L66S mutant toxin from Helicobacter pylori | | Descriptor: | BROMIDE ION, Uncharacterized protein, Toxin | | Authors: | Pathak, C.C, Im, H, Lee, B.J, Yoon, H.J. | | Deposit date: | 2013-07-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
2F25
 
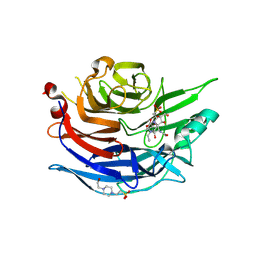 | | Crystal Structure of the Human Sialidase Neu2 E111Q Mutant in Complex with DANA Inhibitor | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 E111Q Mutant in Complex with DANA Inhibitor
To be Published
|
|
4LYC
 
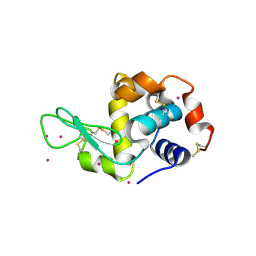 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
2F30
 
 | | Triclinic cross-linked Lysozyme soaked with 4.5M urea | | Descriptor: | Lysozyme C, NITRATE ION, UREA | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
4LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN NEAT ACETONITRILE, THEN BACK-SOAKED IN WATER | | Descriptor: | LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
2X5J
 
 | | Crystal structure of the Apoform of the D-Erythrose-4-phosphate dehydrogenase from E. coli | | Descriptor: | D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, PHOSPHATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2F38
 
 | | Crystal structure of prostaglandin F synathase containing bimatoprost | | Descriptor: | (5Z)-7-{(1R,2R,3R,5S)-3,5-DIHYDROXY-2-[(1E,3S)-3-HYDROXY-5-PHENYLPENT-1-ENYL]CYCLOPENTYL}-N-ETHYLHEPT-5-ENAMIDE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Komoto, J, Yamada, T, Watanabe, K, Woodward, D.F, Takusagawa, F. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prostaglandin F2alpha formation from prostaglandin H2 by prostaglandin F synthase (PGFS): crystal structure of PGFS containing bimatoprost.
Biochemistry, 45, 2006
|
|
2B6T
 
 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural rigidity of a large cavity-containing protein revealed by high-pressure crystallography.
J.Mol.Biol., 367, 2007
|
|
1ZKQ
 
 | | Crystal structure of mouse thioredoxin reductase type 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 2, mitochondrial | | Authors: | Biterova, E.I, Turanov, A.A, Gladyshev, V.N, Barycki, J.J. | | Deposit date: | 2005-05-03 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6KWG
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
2B74
 
 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3K84
 
 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
2F4A
 
 | | Triclinic cross-linked lysozyme soaked with thiourea 1.5M | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION, ... | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-23 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
3FI9
 
 | | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis | | Descriptor: | Malate dehydrogenase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis
To be Published
|
|
6KXL
 
 | | Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-METHOXYETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
4M29
 
 | | Structure of a GH39 Beta-xylosidase from Caulobacter crescentus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-xylosidase | | Authors: | Polo, C.C, Santos, C.R, Correa, J.M, Simao, R.C.G, Seixas, F.A.V, Murakami, M.T. | | Deposit date: | 2013-08-05 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a GH39 Beta-xylosidase from Caulobacter crescentus
Thesis
|
|
4DUV
 
 | | E. coli (lacZ) beta-galactosidase (G974A) 2-deoxy-galactosyl-enzyme and bis-Tris complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-alpha-D-galactopyranose, Beta-galactosidase, ... | | Authors: | Wheatley, R.W, Lo, S, Janzcewicz, L.J, Dugdale, M.L, Huber, R.E. | | Deposit date: | 2012-02-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Glucose Acceptor site of lacZ beta-galactosidase for the synthesis of allolactose - the natural inducer of the lac operon
To be Published
|
|
2WHJ
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
3FJN
 
 | |
3FKE
 
 | | Structure of the Ebola VP35 Interferon Inhibitory Domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Amarasinghe, G.K, Leung, D.W, Ginder, N.D, Honzatko, R.B, Nix, J, Basler, C.F, Fulton, D.B. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Ebola VP35 interferon inhibitory domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QOF
 
 | | Crystal structure of fmn quinone reductase 2 AT 1.55A | | Descriptor: | FLAVIN MONONUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of FMN quinone reductase 2 at 1.55A
To be Published
|
|
2FGN
 
 | |
6AZV
 
 | | IDO1/BMS-978587 crystal structure | | Descriptor: | (1R,2S)-2-(4-[bis(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lewis, H.A. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
