5IFL
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5A6V
 
 | | Open and closed conformations and protonation states of Candida antarctica Lipase B: Xenon complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, LIPASE B, ... | | Authors: | Stauch, B, Fisher, S.J, Cianci, M. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Open and Closed States of Candida Antarctica Lipase B: Protonation and the Mechanism of Interfacial Activation.
J.Lipid Res., 56, 2015
|
|
5A7M
 
 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
5MMI
 
 | | Structure of the large subunit of the chloroplast ribosome | | Descriptor: | 23S ribosomal RNA, 4.5S ribosomal RNA, 50S ribosomal protein 6, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
6CCB
 
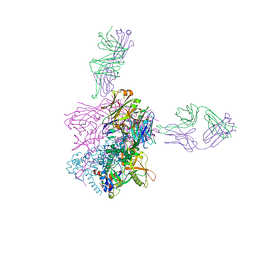 | | Crystal structure of 253-11 SOSIP trimer in complex with 10-1074 Fab | | Descriptor: | 10-1074 FAB heavy chain, 10-1074 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moyo, T, Ereno-Orbea, J, Dorfman, J, Julien, J.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Molecular Basis of Unusually High Neutralization Resistance in Tier 3 HIV-1 Strain 253-11.
J. Virol., 92, 2018
|
|
6K7Y
 
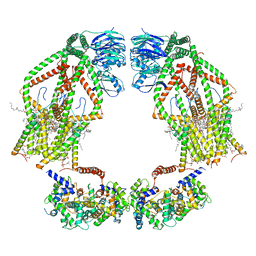 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
5MDZ
 
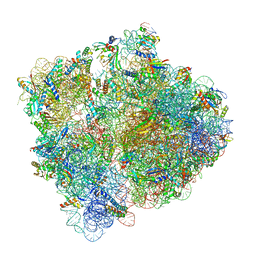 | | Structure of the 70S ribosome (empty A site) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
6DIO
 
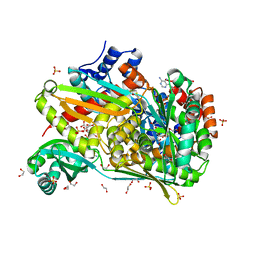 | | Structure of class II HMG-CoA reductase from Delftia acidovorans with NAD bound | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-3-methylglutaryl coenzyme A reductase, CITRIC ACID, ... | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
6KMX
 
 | | Structure of PSI from H. hongdechloris grown under far-red light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
5IRO
 
 | |
4UJD
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
5F0J
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5OMX
 
 | |
5F15
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5OQM
 
 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
6T7T
 
 | | Structure of yeast 80S ribosome stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-23 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6HG5
 
 | | Influenza A virus N6 neuraminidase complex with Oseltamivir (Duck/England/56). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
5IDL
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
5A59
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
6TGE
 
 | | NADP dependent methylene-tetrahydromethanopterin dehydrogenase-NADP+-methenyl-H4MPT+ complex | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional NADP-dependent methylenetetrahydromethanopterin dehydrogenase/methylenetetrahydrofolate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ermler, U, Shima, S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
4UM9
 
 | | Crystal structure of alpha V beta 6 with peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinants of Integrin Beta-Subunit Specificity for Latent Tgf-Beta
Nat.Struct.Mol.Biol., 21, 2014
|
|
6HYT
 
 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.3 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
5IB7
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet, near-cognate tRNALys with U-G mismatch in the A-site and antibiotic paromomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
5IES
 
 | |
