4LEK
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethoxy-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
4LF3
 
 | |
4LEH
 
 | |
4EPS
 
 | |
4LAG
 
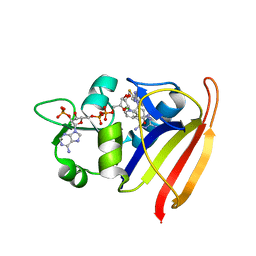 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 6-chloro-7-[5,6-dimethyl-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
3SEE
 
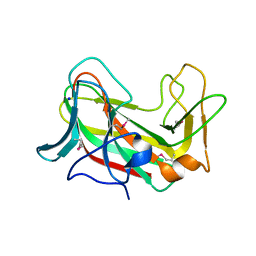 | |
4EIJ
 
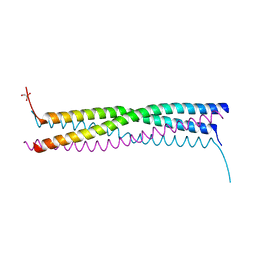 | |
4RH3
 
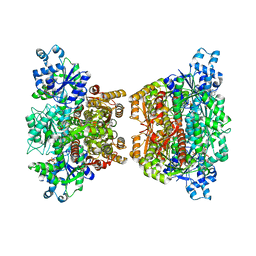 | |
3S25
 
 | |
4EWX
 
 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-04-28 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
4EXX
 
 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-05-01 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
4RJV
 
 | | Crystal Structure of a De Novo Designed Ferredoxin Fold, Northeast Structural Genomics Consortium (NESG) Target OR461 | | Descriptor: | OR461 | | Authors: | O'Connell, P.T, Lin, Y.-R, Guan, R, Koga, N, Koga, R, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR461
To be published
|
|
4L8O
 
 | |
4ROJ
 
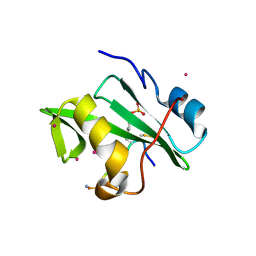 | | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide | | Descriptor: | Guanine nucleotide exchange factor VAV2, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the VAV2 SH2 domain in complex with TXNIP phosphorylated peptide
TO BE PUBLISHED
|
|
4L9R
 
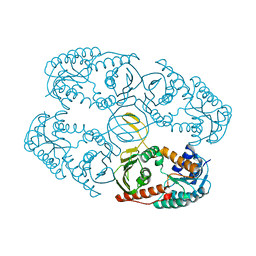 | |
4RPJ
 
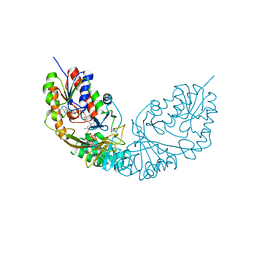 | |
4E6R
 
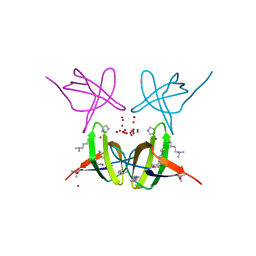 | |
4RKP
 
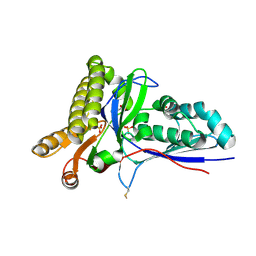 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (apo form) | | Descriptor: | ACETATE ION, Putative uncharacterized protein Ta1305, SULFATE ION | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4L3U
 
 | |
3SCY
 
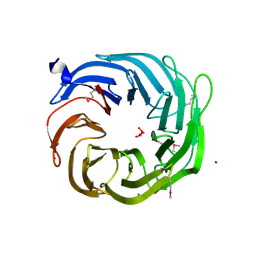 | |
4RZT
 
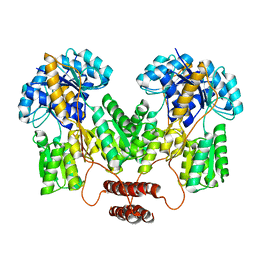 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
4EY1
 
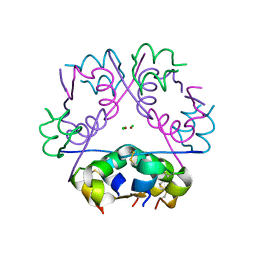 | | Human Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Favero-Retto, M.P, Palmieri, L.C, Lima, L.M.T.R. | | Deposit date: | 2012-05-01 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structural meta-analysis of regular human insulin in pharmaceutical formulations.
Eur J Pharm Biopharm, 85, 2013
|
|
4EIU
 
 | |
4Q9U
 
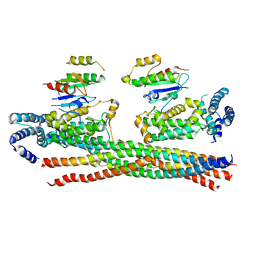 | | Crystal structure of the Rab5, Rabex-5delta and Rabaptin-5C21 complex | | Descriptor: | Rab GTPase-binding effector protein 1, Rab5 GDP/GTP exchange factor, Ras-related protein Rab-5A | | Authors: | Zhang, Z, Zhang, T, Ding, J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.618 Å) | | Cite: | Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5
Elife, 3, 2014
|
|
4QAG
 
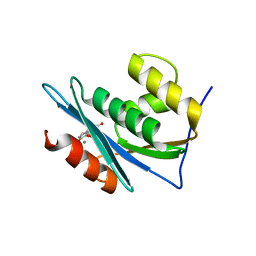 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
