7H8T
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z367659362 (CHIKV_MacB-x1299) | | Descriptor: | 1-(propan-2-yl)-1H-1,3-benzimidazol-2-amine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
5XUQ
 
 | | Crystal structure of VDR-LBD complexed with an antagonist, 2-methylidene-19,26,27-trinor-22-(S)-butyl-1-hydroxy-25-oxo-25-(1H-pyrrol-2-yl)- vitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(1~{H}-pyrrol-2-yl)octan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent Vitamin D Receptor Antagonist
To Be Published
|
|
7H6S
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1162778919 (CHIKV_MacB-x0316) | | Descriptor: | 1-[(5-ethyl-1,2,4-oxadiazol-3-yl)methyl]pyridin-2(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
4O7P
 
 | | Crystal structure of Mycobacterium tuberculosis maltose kinase MaK complexed with maltose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltokinase, SULFATE ION, ... | | Authors: | Li, J, Guan, X.T, Rao, Z.H. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Homotypic dimerization of a maltose kinase for molecular scaffolding.
Sci Rep, 4, 2014
|
|
4OFT
 
 | | C- Orthorombic NaGST1 | | Descriptor: | Glutathione S-transferase-1 | | Authors: | Asojo, O.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of glutathione S-transferase 1 from the major human hookworm parasite Necator americanus (Na-GST-1) in complex with glutathione.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
2A0N
 
 | |
2WC4
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-thia-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
3F07
 
 | | Crystal Structure Analysis of Human HDAC8 complexed with APHA in a new monoclinic crystal form | | Descriptor: | (2E)-N-hydroxy-3-[1-methyl-4-(phenylacetyl)-1H-pyrrol-2-yl]prop-2-enamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3F2U
 
 | | Crystal structure of human chromobox homolog 1 (CBX1) | | Descriptor: | Chromobox protein homolog 1 | | Authors: | Amaya, M.F, Ravichandran, M, Tempel, W, Wernimont, A.K, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Botchkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of human chromobox homolog 1 (CBX1)
To be Published
|
|
3F41
 
 | | Structure of the tandemly repeated protein tyrosine phosphatase like phytase from Mitsuokella multacida | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phytase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a multifunctional, tandemly repeated inositol polyphosphatase.
J.Mol.Biol., 392, 2009
|
|
3VJT
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(R)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
5QJ7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z32327641 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJL
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z56983806 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-N-(3-methylphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3V7Y
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-N-Propargylamino-5'-deoxyadenosine | | Descriptor: | 5'-deoxy-5'-(prop-2-yn-1-ylamino)adenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3A29
 
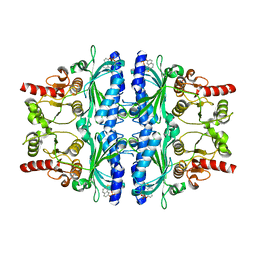 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor | | Descriptor: | 2-amino-4,5-dihydronaphtho[1,2-d][1,3]thiazol-8-yl dihydrogen phosphate, Fructose-1,6-bisphosphatase 1 | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR, and X-ray structure of tricyclic compounds as potent FBPase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4JDC
 
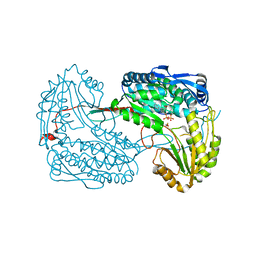 | |
4OH3
 
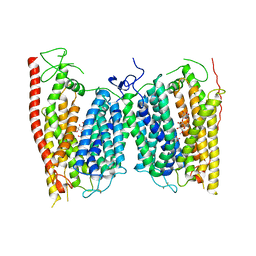 | | Crystal structure of a nitrate transporter | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, NITRATE ION, Nitrate transporter 1.1 | | Authors: | Sun, J, Bankston, J.R, Payandeh, J, Hinds, T.R, Zagotta, W.N, Zheng, N. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the plant dual-affinity nitrate transporter NRT1.1.
Nature, 507, 2014
|
|
7HIN
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188476-02 (CHIKV_MacB-x2343) | | Descriptor: | 1-({5-[(1R)-1-(4-bromo-3-fluorophenyl)ethyl]-1,2,4-oxadiazol-3-yl}methyl)pyridin-2(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
7HIL
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188488-01 (CHIKV_MacB-x2129) | | Descriptor: | 1-ethyl-N-methyl-6-oxo-N-[(1,3-thiazol-2-yl)methyl]-1,6-dihydropyridazine-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
2DMT
 
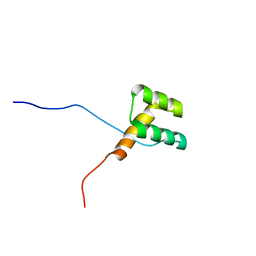 | | Solution structure of the homeobox domain of Homeobox protein BarH-like 1 | | Descriptor: | Homeobox protein BarH-like 1 | | Authors: | Ohnishi, S, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of Homeobox protein BarH-like 1
To be Published
|
|
4J70
 
 | | Yeast 20S proteasome in complex with the belactosin derivative 3e | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Kawamura, S, Unno, Y, List, A, Tanaka, M, Sasaki, T, Arisawa, M, Asai, A, Groll, M, Shuto, S. | | Deposit date: | 2013-02-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent Proteasome Inhibitors Derived from the Unnatural cis-Cyclopropane Isomer of Belactosin A: Synthesis, Biological Activity, and Mode of Action.
J.Med.Chem., 56, 2013
|
|
4GDX
 
 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
7HIU
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188492-02 (CHIKV_MacB-x2399) | | Descriptor: | 1-({5-[(1S)-1-(4-methoxyphenyl)ethyl]-1,2,4-oxadiazol-3-yl}methyl)pyridin-2(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
3LP4
 
 | |
1HL7
 
 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
