7XZO
 
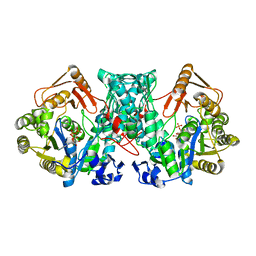 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
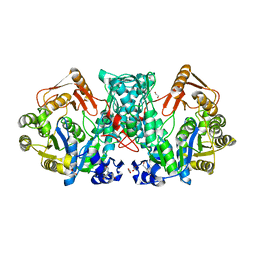
7XZN
 
 | |
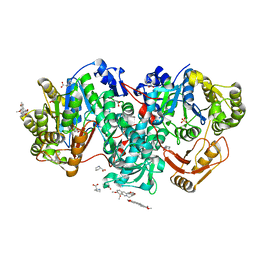
7XZP
 
 | |
6GJE
 
 | | Structure of the Amnionless(20-357)-Cubilin(36-135) complex | | Descriptor: | Cubilin, Protein amnionless | | Authors: | Larsen, C, Etzerodt, A, Madsen, M, Skjoedt, K, Moestrup, S.K, Andersen, C.B.F. | | Deposit date: | 2018-05-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assembly of the megadalton-sized receptor for intestinal vitamin B12uptake and kidney protein reabsorption.
Nat Commun, 9, 2018
|
|
7ZTH
 
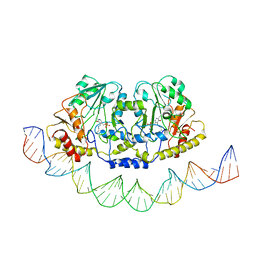 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
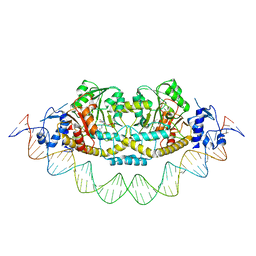 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
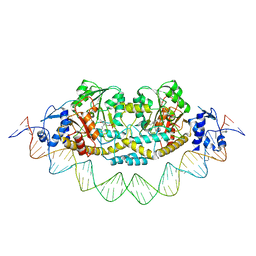 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
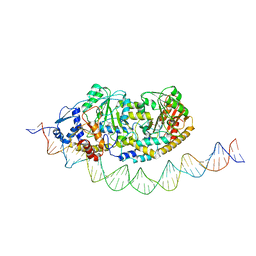 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
8BTR
 
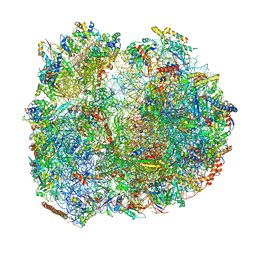 | | Giardia Ribosome in PRE-T Hybrid State (D2) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BTD
 
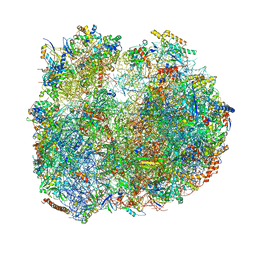 | | Giardia Ribosome in PRE-T Hybrid State (D1) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-28 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BR8
 
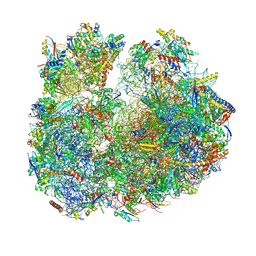 | | Giardia ribosome in POST-T state (A1) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BRM
 
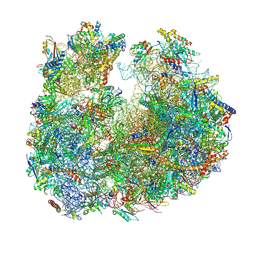 | | Giardia ribosome in POST-T state, no E-site tRNA (A6) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-23 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
6HGL
 
 | |
7VJU
 
 | |
1HXH
 
 | | COMAMONAS TESTOSTERONI 3BETA/17BETA HYDROXYSTEROID DEHYDROGENASE | | Descriptor: | 3BETA/17BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Benach, J, Filling, C, Oppermann, U.C.T, Roversi, P, Bricogne, G, Berndt, K.D, Jornvall, H, Ladenstein, R. | | Deposit date: | 2001-01-15 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of Bacterial 3beta/17beta-Hydroxysteroid Dehydrogenase at 1.2 A Resolution: A Model for
Multiple Steroid Recognition
Biochemistry, 41, 2002
|
|
6ZE0
 
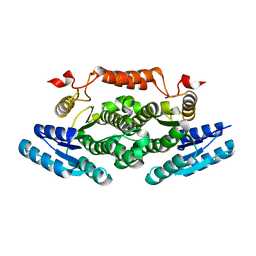 | |
6ZDZ
 
 | |
1OCV
 
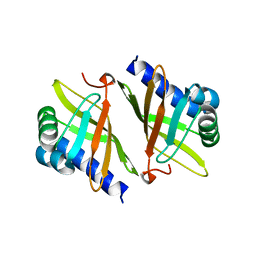 | |
6LY4
 
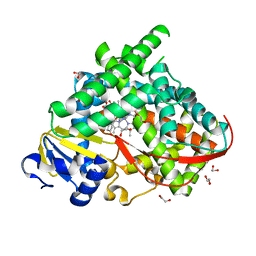 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
9EQG
 
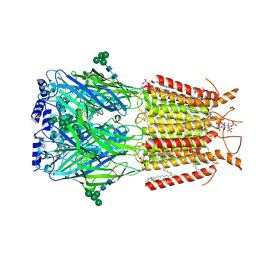 | |
7KGY
 
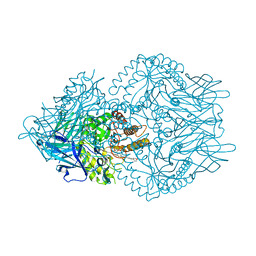 | | Beta-glucuronidase from Faecalibacterium prausnitzii bound to the inhibitor UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microbial enzymes induce colitis by reactivating triclosan in the mouse gastrointestinal tract.
Nat Commun, 13, 2022
|
|
7KGZ
 
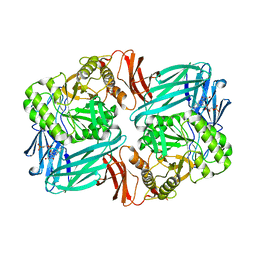 | |
7WZD
 
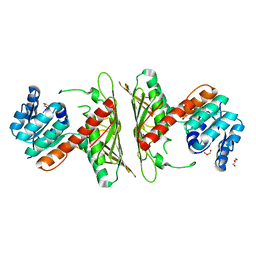 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase from Comamonas testosteroni KF1 | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7Y98
 
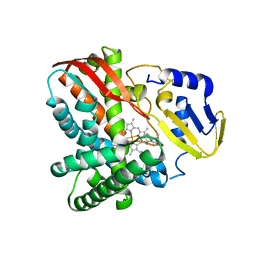 | | Crystal structure of CYP109B4 from Bacillus Sonorensis in complex with Testosterone | | Descriptor: | Cytochrome P450 monooxygenase YjiB, PROTOPORPHYRIN IX CONTAINING FE, TESTOSTERONE | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
3AKM
 
 | | X-ray structure of iFABP from human and rat with bound fluorescent fatty acid analogue | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, Fatty acid-binding protein, intestinal, ... | | Authors: | Wielens, J, Laguerre, A.J.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-07-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human and rat intestinal fatty acid binding proteins in complex with 11-(Dansylamino)undecanoic acid
To be Published
|
|
