7E5P
 
 | | Aptamer enhancing peroxidase activity of myoglobin | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*AP*GP*GP*G)-3') | | Authors: | Tsukakoshi, K, Matsugami, A, Khunathai, K, Kanazashi, M, Yamagishi, Y, Nakama, K, Oshikawa, D, Hayashi, F, Kuno, H, Ikebukuro, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol.
Nucleic Acids Res., 49, 2021
|
|
7EQD
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLUM RUBRUM | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Yu, L.-J, Hall, M, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-05-01 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM Structure of the Photosynthetic LH1-RC Complex from Rhodospirillum rubrum .
Biochemistry, 2021
|
|
7EY7
 
 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EXT
 
 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
2NOH
 
 | | Structure of catalytically inactive Q315A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOZ
 
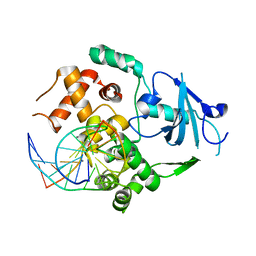 | | Structure of Q315F human 8-oxoguanine glycosylase distal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*G*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
7EXZ
 
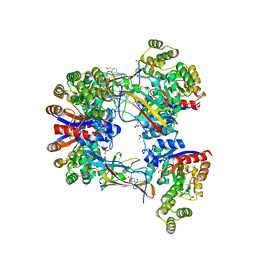 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
2NOF
 
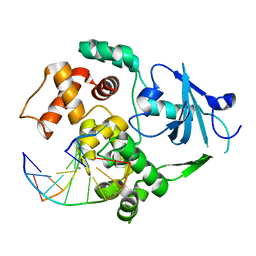 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOI
 
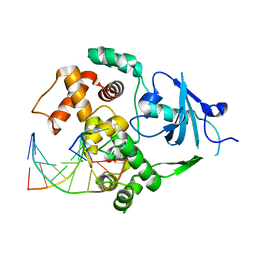 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOB
 
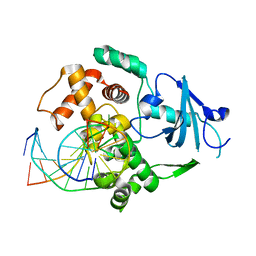 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
7F0L
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES MONOMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein beta chain, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Nagashima, V.P, Kanno, R, Kawamura, S, Kikuchi, R, Ji, X.-C, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex.
Nat Commun, 12, 2021
|
|
7FEM
 
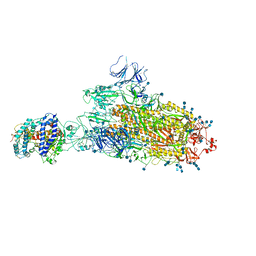 | | SARS-CoV-2 B.1.1.7 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wen, Z.L, Zhu, Y, Sun, F. | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure-based evidence for the enhanced transmissibility of the dominant SARS-CoV-2 B.1.1.7 variant (Alpha).
Cell Discov, 7, 2021
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7F36
 
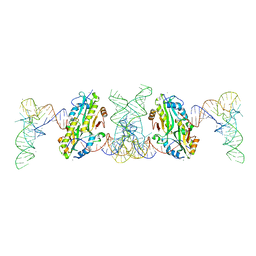 | | TacT complexed with acetyl-glycyl-tRNAGly | | Descriptor: | ACETYLAMINO-ACETIC ACID, CALCIUM ION, N-acetyltransferase domain-containing protein, ... | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Molecular basis of glycyl-tRNAGly acetylation by TacT from Salmonella Typhimurium
Cell Rep, 37, 2021
|
|
7FJL
 
 | |
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | Authors: | Zhang, L, Wang, X, Shan, S, Zhang, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7EW6
 
 | | Barley photosystem I-LHCI-Lhca5 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-24 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7F9O
 
 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EWK
 
 | | Barley photosystem I-LHCI-Lhca6 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7FCE
 
 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (closed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2NMU
 
 | | Crystal structure of the hypothetical protein from Salmonella typhimurium LT2. Northeast Structural Genomics Consortium target StR127. | | Descriptor: | Putative DNA-binding protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical protein from Salmonella typhimurium LT2. Northeast Structural Genomics Consortium target StR127.
To be Published
|
|
2NOE
 
 | | Structure of catalytically inactive G42A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2N4P
 
 | | Solution structure of the n-terminal domain of tdp-43 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F, Buratti, E, Laurents, D.V. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The TDP-43 N-terminal domain structure at high resolution.
Febs J., 283, 2016
|
|
