5SPY
 
 | |
1SAN
 
 | |
3SJT
 
 | | Crystal structure of human arginase I in complex with the inhibitor Me-ABH, Resolution 1.60 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxyhexyl](trihydroxy)borate | | Authors: | Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
3SMR
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3MVH
 
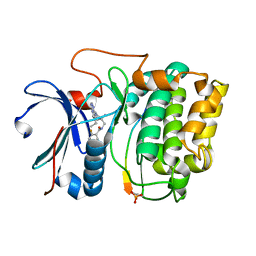 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | GSK3-beta peptide, MANGANESE (II) ION, N-{[(3S)-3-amino-1-(5-ethyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-yl]methyl}-2,4-difluorobenzamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
4HRC
 
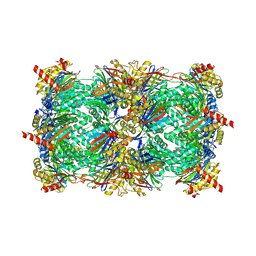 | | Crystal structure of yeast 20S proteasome in complex with epoxyketone carmaphycin analogue 3 | | Descriptor: | N-hexanoyl-L-valyl-N~1~-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-N~5~,N~5~-dimethyl-L-glutamamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-27 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
3TWT
 
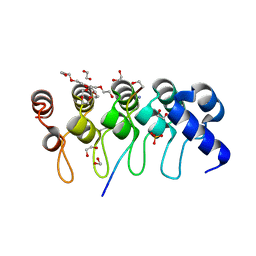 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human MCL1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, NONAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
2BOP
 
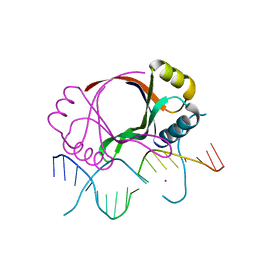 | | CRYSTAL STRUCTURE AT 1.7 ANGSTROMS OF THE BOVINE PAPILLOMAVIRUS-1 E2 DNA-BINDING DOMAIN BOUND TO ITS DNA TARGET | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*TP*CP*G )-3'), PROTEIN (E2), YTTERBIUM (III) ION | | Authors: | Hegde, R.S, Grossman, S.R, Laimins, L.A, Sigler, P.B. | | Deposit date: | 1994-01-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target.
Nature, 359, 1992
|
|
3N0X
 
 | |
3T1L
 
 | |
1VP7
 
 | |
5SSL
 
 | |
5SS6
 
 | |
4BSX
 
 | | Crystal Structure of the N terminal domain of TRIF (TIR-domain- containing adapter-inducing interferon-beta) | | Descriptor: | TIR DOMAIN-CONTAINING ADAPTER MOLECULE 1 | | Authors: | Ullah, M.O, Ve, T, Mangan, M, Alaidarous, M, Sweet, M.J, Mansell, A, Kobe, B. | | Deposit date: | 2013-06-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | The Tlr Signalling Adaptor Trif/Ticam-1 Has an N-Terminal Helical Domain with Structural Similarity to Ifit Proteins
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1RWY
 
 | | CRYSTAL STRUCTURE OF RAT ALPHA-PARVALBUMIN AT 1.05 RESOLUTION | | Descriptor: | ACETIC ACID, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Bottoms, C.A, Schuermann, J.P, Agah, S, Henzl, M.T, Tanner, J.J. | | Deposit date: | 2003-12-17 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Structure of Rat Alpha-Parvalbumin at 1.05 Resolution
Protein Sci., 13, 2004
|
|
4CDF
 
 | | Human DPP1 in complex with (2S,4S)-N-((1S)-1-cyano-2-(4-(4- cyanophenyl)phenyl)ethyl)-4-hydroxy-piperidine-2-carboxamide | | Descriptor: | (2S,4S)-N-[(2S)-1-azanylidene-3-[4-(4-cyanophenyl)phenyl]propan-2-yl]-4-oxidanyl-piperidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Debreczeni, J, Edman, K, Furber, M, Tiden, A, Gardiner, P, Mete, T, Ford, R, Millichip, I, Stein, L, Mather, A, Kinchin, E, Luckhurst, C, Cage, P, Sanghanee, H, Breed, J, Wissler, L. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cathepsin C Inhibitors: Property Optimization and Identification of a Clinical Candidate.
J.Med.Chem., 57, 2014
|
|
3RIE
 
 | | The structure of Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Williams, M, Sprenger, J, Persson, L, Louw, A.I, Birkholtz, L, Al-Karadaghi, S. | | Deposit date: | 2011-04-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel inhibitor of Plasmodium falciparum spermidine synthase: a twist in the tail.
Malar J, 14, 2015
|
|
1OC6
 
 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
5SDV
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(c(C(=O)NCc1cocn1)cnn2C)C(=O)Nc4nc3nc(cn3cc4)c5ccccc5, micromolar IC50=0.001258 | | Descriptor: | 1-methyl-N~4~-[(1,3-oxazol-4-yl)methyl]-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-4,5-dicarboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7K4H
 
 | | Human Arginase 1 in complex with compound 04. | | Descriptor: | 3-[(1~{R},5~{S},8~{R})-5-carboxy-2,6-diazabicyclo[3.2.1]octan-8-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1O54
 
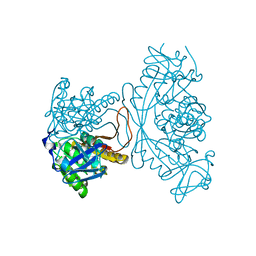 | |
6HGB
 
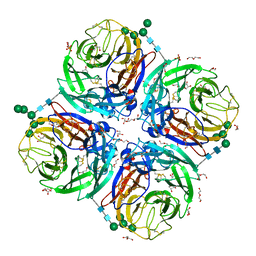 | | Influenza A virus N6 neuraminidase native structure (Duck/England/56). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-23 | | Release date: | 2018-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
7K4J
 
 | | Human Arginase 1 in complex with compound 51. | | Descriptor: | 3-[(1~{S},2~{S},5~{R})-2-carboxy-3,6-diazabicyclo[3.2.0]heptan-1-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
2VTC
 
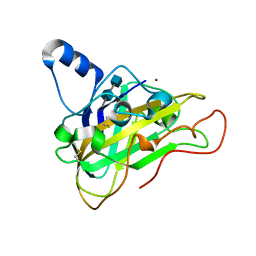 | | The structure of a glycoside hydrolase family 61 member, Cel61B from the Hypocrea jecorina. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEL61B, NICKEL (II) ION | | Authors: | Karkehabadi, S, Hansson, H, Kim, S, Piens, K, Mitchinson, C, Sandgren, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The First Structure of a Glycoside Hydrolase Family 61 Member, Cel61B from the Hypocrea Jecorina, at 1.6 A Resolution.
J.Mol.Biol., 383, 2008
|
|
1VRA
 
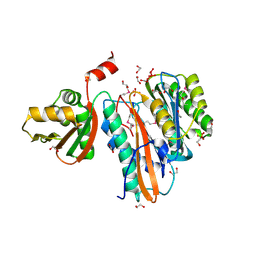 | |
