1HTH
 
 | | The solution structure of cyclic human parathyroid hormone fragment 1-34, NMR, 10 structures | | Descriptor: | CYCLIC PARATHYROID HORMONE | | Authors: | Roesch, P, Seidel, G, Schaefer, W, Esswein, A, Hofmann, E. | | Deposit date: | 1997-04-09 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The structure of human parathyroid hormone-related protein(1-34) in near-physiological solution.
Febs Lett., 444, 1999
|
|
3D9G
 
 | | Nitroalkane oxidase: wild type crystallized in a trapped state forming a cyanoadduct with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Nitroalkane oxidase, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
7AWE
 
 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [(1R)-2-(1-benzofuran-3-yl)-1-{[(1S,2R,4R)-7-oxabicyclo[2.2.1]heptan-2-yl]formamido}ethyl]boronic acid | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Musil, D, Klein, M. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i) Delivering Efficacy in Multiple Myeloma Models.
Mol.Cancer Ther., 20, 2021
|
|
1AFK
 
 | |
1OWP
 
 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2OKX
 
 | | Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A | | Descriptor: | CALCIUM ION, GLYCEROL, Rhamnosidase B | | Authors: | Cui, Z, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 78 alpha-L-Rhamnosidase from Bacillus sp. GL1
J.Mol.Biol., 374, 2007
|
|
5EHQ
 
 | | mAChE-anti TZ2PA5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[1-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
4LEL
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCG-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.90000033 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WQP
 
 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | Deposit date: | 2014-01-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
2XH4
 
 | |
3MCX
 
 | |
2XH1
 
 | | Crystal structure of human KAT II-inhibitor complex | | Descriptor: | (3S)-10-(4-AMINOPIPERAZIN-1-YL)-9-FLUORO-7-HYDROXY-3-METHYL-2,3-DIHYDRO-8H-[1,4]OXAZINO[2,3,4-IJ]QUINOLINE-6-CARBOXYLATE, GLYCEROL, IODIDE ION, ... | | Authors: | Rossi, F, Casazza, V, Garavaglia, S, Sathyasaikumar, K.V, Schwarcz, R, Kojima, S.I, Okuwaki, K, Ono, S.I, Kajii, Y, Rizzi, M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure-Based Selective Targeting of the Pyridoxal 5'-Phsosphate Dependent Enzyme Kynurenine Aminotransferase II for Cognitive Enhancement
J.Med.Chem., 53, 2010
|
|
4PJ9
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT TRAJ20 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
4L97
 
 | | Structure of the RBP of lactococcal phage 1358 in complex with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
1OX0
 
 | |
6CCL
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 1-benzyl-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1-benzyl-1H-imidazo[4,5-b]pyridine, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
3R68
 
 | | Molecular Analysis of the PDZ3 domain of PDZK1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kocher, O, Birrane, G, Krieger, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of the PDZ3 Domain of the Adaptor Protein PDZK1 as a Second, Physiologically Functional Binding Site for the C Terminus of the High Density Lipoprotein Receptor Scavenger Receptor Class B Type I.
J.Biol.Chem., 286, 2011
|
|
1OWL
 
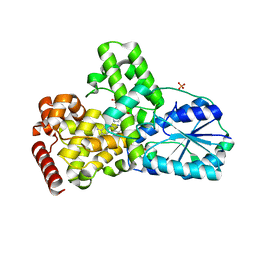 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2XH7
 
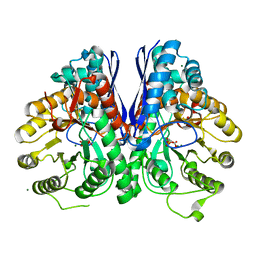 | |
3DX1
 
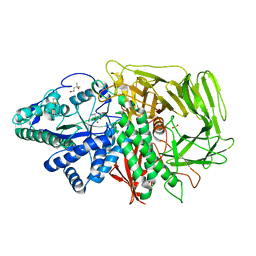 | | Golgi alpha-Mannosidase II in complex with Mannostatin analog (1S,2S,3R,4R)-4-aminocyclopentane-1,2,3-triol | | Descriptor: | (1S,2S,3R,4R)-4-aminocyclopentane-1,2,3-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
1LD7
 
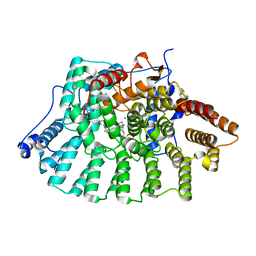 | | Co-crystal structure of Human Farnesyltransferase with farnesyldiphosphate and inhibitor compound 66 | | Descriptor: | (20S)-19,20,22,23-TETRAHYDRO-19-OXO-5H,21H-18,20-ETHANO-12,14-ETHENO-6,10-METHENOBENZ[D]IMIDAZO[4,3-L][1,6,9,13]OXATRIA ZACYCLONOADECOSINE-9-CARBONITRILE, FARNESYL DIPHOSPHATE, ZINC ION, ... | | Authors: | Taylor, J.S, Terry, K.L, Beese, L.S. | | Deposit date: | 2002-04-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Aminopyrrolidinone farnesyltransferase inhibitors: design of macrocyclic compounds with improved pharmacokinetics and excellent cell potency.
J.Med.Chem., 45, 2002
|
|
7JNW
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)isobutyramide | | Descriptor: | 2-methyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
3BLN
 
 | |
7JNV
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butyramide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
3DRO
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN grown in ammonium sulfate | | Descriptor: | 2F5 Fab heavy chain, 2F5 Fab light chain, ELLELDKWASLWN gp41 peptide | | Authors: | Julien, J.-P, Bryson, S, Pai, E.F. | | Deposit date: | 2008-07-11 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
