3IU7
 
 | |
1U13
 
 | |
3IU9
 
 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T07 | | Descriptor: | 5-[(2,4-dichlorobenzyl)sulfanyl]-4H-1,2,4-triazol-3-amine, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
3IU8
 
 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T03 | | Descriptor: | 3-[(4-fluorobenzyl)sulfanyl]-4H-1,2,4-triazole, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
1XKF
 
 | |
3G5F
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2IJ5
 
 | |
3G5H
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Y5H
 
 | |
1SM8
 
 | | M. tuberculosis dUTPase complexed with chromium and dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHROMIUM ION, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
2XJA
 
 | | Structure of MurE from M.tuberculosis with dipeptide and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Basavannacharya, C, Moody, P.R, Bhakta, S, Keep, N. | | Deposit date: | 2010-07-03 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Essential Residues for the Enzyme Activity of ATP-Dependent Mure Ligase from Mycobacterium Tuberculosis.
Protein Cell, 1, 2010
|
|
4KMC
 
 | |
1MC9
 
 | | STREPROMYCES LIVIDANS XYLAN BINDING DOMAIN CBM13 IN COMPLEX WITH XYLOPENTAOSE | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, SULFATE ION, ... | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2002-08-06 | | Release date: | 2002-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
3C8N
 
 | |
4FQD
 
 | |
3H7Q
 
 | |
2G85
 
 | | Crystal structure of chorismate synthase from Mycobacterium tuberculosis at 2.22 angstrons of resolution | | Descriptor: | Chorismate synthase | | Authors: | Dias, M.V.B, dos Santos, B.B, Ely, F, Basso, L.A, Santos, D.S, de Azevedo Jr, W.F. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of chorismate synthase from Mycobacterium tuberculosis at 2.22 angstron of resolution
To be Published
|
|
2AP9
 
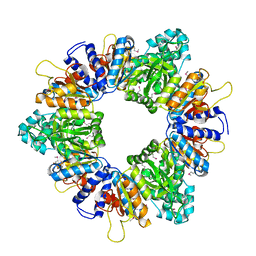 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
1ZAU
 
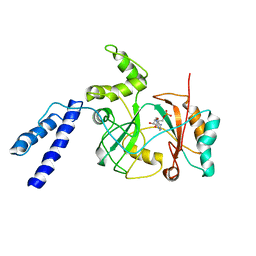 | |
6WF6
 
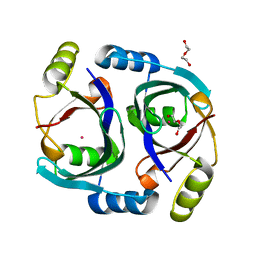 | | Streptomyces coelicolor methylmalonyl-CoA epimerase | | Descriptor: | COBALT (II) ION, DI(HYDROXYETHYL)ETHER, Methylmalonyl-CoA epimerase | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
6WFH
 
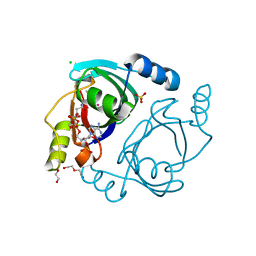 | | Streptomyces coelicolor methylmalonyl-CoA epimerase substrate complex | | Descriptor: | (3S,5R,9R,19E)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,19-tetrahydroxy-8,8,20-trimethyl-10,14-dioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphahenicos-19-en-21-oic acid 3,5-dioxide (non-preferred name), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
1QFI
 
 | | SYNTHESIS AND STRUCTURE OF PROLINE RING MODIFIED ACTINOMYCINS OF X TYPE | | Descriptor: | ACTINOMYCIN X2, ETHYL ACETATE, METHANOL | | Authors: | Lifferth, A, Bahner, I, Lackner, H, Schaefer, M. | | Deposit date: | 1999-04-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Synthesis and Structure of Proline Ring Modified Actinomycins of the X-Type
Z.Naturforsch., 54, 1999
|
|
2XB8
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase in complex with inhibitor compound (2R)-2-(4-methoxybenzyl)-3- dehydroquinic acid | | Descriptor: | (1R,2R,4S,5R)-1,4,5-TRIHYDROXY-2-(4-METHOXYBENZYL)-3-OXOCYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Tizon, L, Llamas-Saiz, A.L, Fox, G.C, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the Key Factors that Control the Inhibition of Type II Dehydroquinase by (2R)-2- Benzyl-3-Dehydroquinic Acids.
Chemmedchem, 5, 2010
|
|
4KV3
 
 | |
2CIN
 
 | |
