5EWA
 
 | | Crystal structure of the metallo-beta-lactamase IMP-1 in complex with the bisthiazolidine inhibitor L-VC26 | | Descriptor: | (3~{R},5~{R},7~{a}~{S})-2,2-dimethyl-5-(sulfanylmethyl)-3,5,7,7~{a}-tetrahydro-[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Kosmopoulou, M, Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4HO9
 
 | | Crystal structure of glucose 1-phosphate thymidylyltransferase from Aneurinibacillus thermoaerophilus complexed with UDP-galactose and UTP | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, SULFATE ION, ... | | Authors: | Chen, T.J, Chien, W.T, Lin, C.C, Wang, W.C. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glucose 1-phosphate thymidylyltransferase from Aneurinibacillus thermoaerophilus complexed with UDP-galactose and UTP
TO BE PUBLISHED
|
|
7FQH
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(3S)-5-amino-5-oxopent-1-yn-3-yl]-1-[1-[4-(cyclopropylmethoxy)phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Hewings-David, S, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
4IB2
 
 | |
6J4Z
 
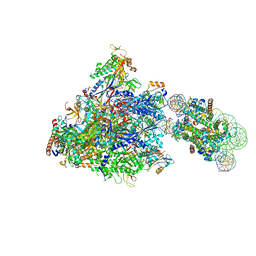 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
1HQN
 
 | | THE SELENOMETHIONINE DERIVATIVE OF P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2000-12-18 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7API
 
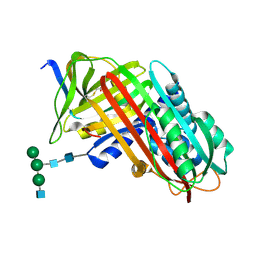 | | THE S VARIANT OF HUMAN ALPHA1-ANTITRYPSIN, STRUCTURE AND IMPLICATIONS FOR FUNCTION AND METABOLISM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTITRYPSIN, ... | | Authors: | Loebermann, H, Tokuoka, R, Deisenhofer, J, Huber, R. | | Deposit date: | 1988-09-08 | | Release date: | 1990-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism.
Protein Eng., 2, 1989
|
|
2NSH
 
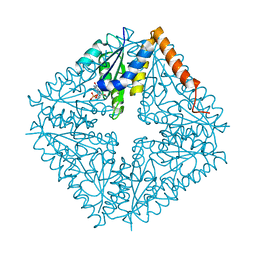 | | E. coli PurE H45Q mutant complexed with nitro-AIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2NSQ
 
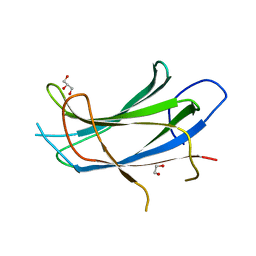 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
5SLY
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1526504764 | | Descriptor: | 1-(1-ethyl-1H-pyrazol-5-yl)-N-methylmethanamine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3B8L
 
 | |
1P7T
 
 | | Structure of Escherichia coli malate synthase G:pyruvate:acetyl-Coenzyme A abortive ternary complex at 1.95 angstrom resolution | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anstrom, D.M, Kallio, K, Remington, S.J. | | Deposit date: | 2003-05-05 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia Coli Malate Synthase G:pyruvate:acetyl-coenzyme A Abortive Ternary Complex at 1.95 Angstrom Resolution
Protein Sci., 12, 2003
|
|
3B00
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with cetyltrimethylammonium bromide | | Descriptor: | CALCIUM ION, CETYL-TRIMETHYL-AMMONIUM, Laminarinase | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
4CAE
 
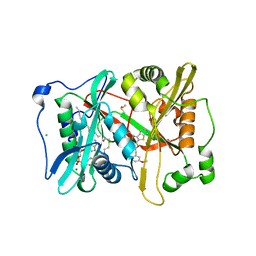 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor (compound 20b) | | Descriptor: | 2-oxopentadecyl-CoA, 4-[(2-{5-[(3-METHOXYPHENYL)METHYL]-1,3,4-OXADIAZOL-2-YL}-1-BENZOTHIOPHEN-3-YL)OXY]PIPERIDINE, CHLORIDE ION, ... | | Authors: | Rackham, M.D, Brannigan, J.A, Rangachari, K, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Design and Synthesis of High Affinity Inhibitors of Plasmodium Falciparum and Plasmodium Vivax N-Myristoyltransferases Directed by Ligand Efficiency Dependent Lipophilicity (Lelp).
J.Med.Chem., 57, 2014
|
|
4PM7
 
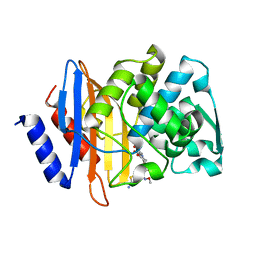 | | Crystal structure of CTX-M-14 S70G:S237A in complex with cefotaxime at 1.29 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PNI
 
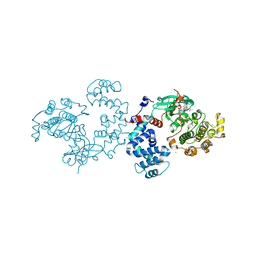 | | Bovine G protein-coupled receptor kinase 1 in complex with GSK2163632A | | Descriptor: | 3-[(2-{[1-(N,N-dimethylglycyl)-6-methoxy-4,4-dimethyl-1,2,3,4-tetrahydroquinolin-7-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]thiophene-2-carboxamide, CHLORIDE ION, Rhodopsin kinase | | Authors: | Homan, K.T, Tesmer, J.J.G. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and structure-function analysis of subfamily selective g protein-coupled receptor kinase inhibitors.
Acs Chem.Biol., 10, 2015
|
|
1OQ5
 
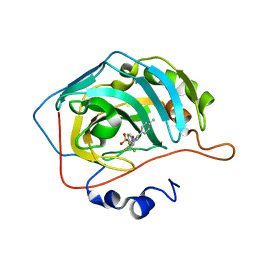 | | CARBONIC ANHYDRASE II IN COMPLEX WITH NANOMOLAR INHIBITOR | | Descriptor: | 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, Carbonic anhydrase II, ZINC ION | | Authors: | Weber, A, Casini, A, Heine, A, Kuhn, D, Supuran, C.T, Scozzafava, A, Klebe, G. | | Deposit date: | 2003-03-07 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected nanomolar inhibition of carbonic anhydrase by COX-2-selective celecoxib: new pharmacological opportunities due to related binding site recognition.
J.Med.Chem., 47, 2004
|
|
4HYW
 
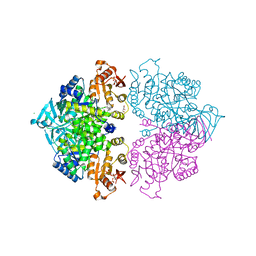 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7K3O
 
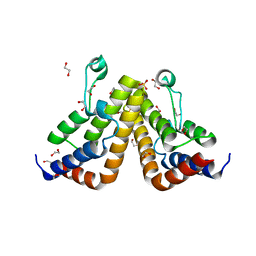 | |
1XIH
 
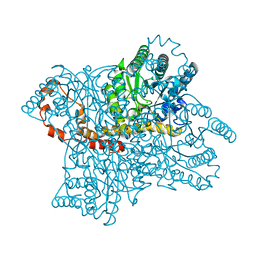 | |
7K0B
 
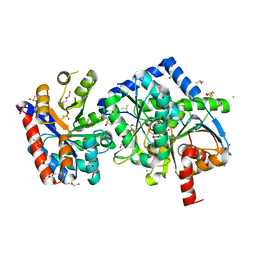 | |
4KTR
 
 | | Crystal structure of 2-O-alpha-glucosylglycerol phosphorylase in complex with isofagomine and glycerol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Touhara, K.K, Nihira, T, Kitaoka, M, Nakai, H, Fushinobu, S. | | Deposit date: | 2013-05-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-alpha-glucosylglycerol phosphorylase
J.Biol.Chem., 289, 2014
|
|
3SNF
 
 | | Onconase, atomic resolution crystal structure | | Descriptor: | ACETATE ION, Protein P-30, SULFATE ION | | Authors: | Holloway, D.E, Singh, U.P, Shogen, K, Acharya, K.R. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of Onconase at 1.1 angstrom resolution--insights into substrate binding and collective motion.
Febs J., 278, 2011
|
|
7G5F
 
 | | Crystal Structure of rat Autotaxin in complex with 1H-benzotriazol-5-yl-[rac-(1R,2R,6S,7S)-9-[4-(cyclopropylmethoxy)naphthalene-2-carbonyl]-4,9-diazatricyclo[5.3.0.02,6]decan-4-yl]methanone, i.e. SMILES N1(C[C@H]2[C@@H](C1)[C@H]1[C@@H]2CN(C1)C(=O)c1ccc2c(c1)N=NN2)C(=O)c1cc(c2c(c1)cccc2)OCC1CC1 with IC50=0.00985223 microM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
1UVA
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | MYRISTIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN 1 | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
