5C0Y
 
 | |
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
5GUH
 
 | | Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA | | Descriptor: | MAGNESIUM ION, PIWI, RNA (28-MER) | | Authors: | Matsumoto, N, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA
Cell, 167, 2016
|
|
2YH1
 
 | | Model of human U2AF65 tandem RRM1 and RRM2 domains with eight-site uridine binding | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP)-3', SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Mackereth, C.D, Madl, T, Simon, B, Zanier, K, Gasch, A, Sattler, M. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-Domain Conformational Selection Underlies Pre-Mrna Splicing Regulation by U2Af
Nature, 475, 2011
|
|
6IS0
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
2IWA
 
 | | Unbound glutaminyl cyclotransferase from Carica papaya. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLUTAMINE CYCLOTRANSFERASE, ... | | Authors: | Guevara, T, Mallorqui-Fernandez, N, Garcia-Castellanos, R, Petersen, G.E, Lauritzen, C, Pedersen, J, Arnau, J, Gomis-Ruth, F.X, Sola, M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Papaya Glutamine Cyclotransferase Shows a Singular Five-Fold Beta-Propeller Architecture that Suggests a Novel Reaction Mechanism.
Biol.Chem., 387, 2006
|
|
6N3D
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif (APO-State) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6N3E
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to U2AF ligand motif 4 | | Descriptor: | FORMIC ACID, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6N3F
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to SF3b1 ULM5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Leach, J.R, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
8JOZ
 
 | |
4MDX
 
 | |
2Y0S
 
 | | Crystal structure of Sulfolobus shibatae RNA polymerase in P21 space group | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, DNA-DIRECTED RNA POLYMERASE SUBUNIT A'', DNA-DIRECTED RNA POLYMERASE SUBUNIT D, ... | | Authors: | Wojtas, M, Peralta, B, Ondiviela, M, Mogni, M, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2010-12-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Archaeal RNA polymerase: the influence of the protruding stalk in crystal packing and preliminary biophysical analysis of the Rpo13 subunit.
Biochem. Soc. Trans., 39, 2011
|
|
4M59
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
6TL0
 
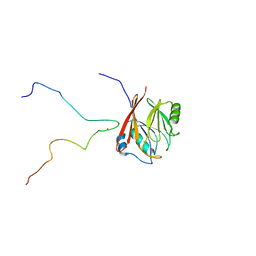 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of VPS29 with VARP 687-747 | | Descriptor: | Ankyrin repeat domain-containing protein 27, Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Crawley-Snowdon, H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Nat Commun, 11, 2020
|
|
3IZ4
 
 | |
8XRJ
 
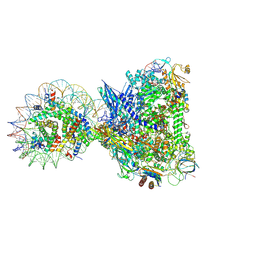 | | RNA polymerase II elongation complex with upstream nucleosome extracted from human nuclei | | Descriptor: | DNA, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Kujirai, T, Kato, J, Yamamoto, K, Hirai, S, Negishi, L, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-01-07 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Multiple structures of RNA polymerase II isolated from human nuclei by ChIP-CryoEM analysis.
Nat Commun, 16, 2025
|
|
8XVS
 
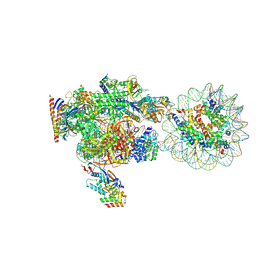 | | RNA polymerase II elongation complex with downstream nucleosome extracted from human nuclei | | Descriptor: | DNA, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Kujirai, T, Kato, J, Yamamoto, K, Hirai, S, Negishi, L, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-01-15 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Multiple structures of RNA polymerase II isolated from human nuclei by ChIP-CryoEM analysis.
Nat Commun, 16, 2025
|
|
3AEV
 
 | | Crystal structure of a/eIF2alpha-aDim2p-rRNA complex from Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH1566, RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*CP*UP*CP*C)-3'), Translation initiation factor 2 subunit alpha | | Authors: | Tanokura, M, Jia, M.Z, Nagata, K. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3' end fragment of 16S rRNA
J.Mol.Biol., 398, 2010
|
|
5I4A
 
 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | Descriptor: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | Authors: | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8B7B
 
 | | Tubulin - maytansinoid - 6 complex | | Descriptor: | (~{Z})-11-[[(1~{R},2~{R},3~{S},5~{S},6~{S},16~{E},18~{E},20~{R},21~{S})-11-chloranyl-12,20-dimethoxy-2,5,9,16-tetramethyl-21-oxidanyl-8,23-bis(oxidanylidene)-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1^{10,14}.0^{3,5}]hexacosa-10(26),11,13,16,18-pentaen-6-yl]oxy]-11-oxidanylidene-undec-2-enoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, J.F, Steinmetz, M.O, Prota, A.E, Pieraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
8B7A
 
 | | Tubulin - maytansinoid - 4 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, F.J, Steinmetz, M.O, Prota, A.E, Piaraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
8B7C
 
 | | Tubulin-maytansinoid-12 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, J.F, Steinmetz, M.O, Prota, A.E, Pieraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
2YH0
 
 | | Solution structure of the closed conformation of human U2AF65 tandem RRM1 and RRM2 domains | | Descriptor: | SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Mackereth, C.D, Madl, T, Simon, B, Zanier, K, Gasch, A, Sattler, M. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-Domain Conformational Selection Underlies Pre-Mrna Splicing Regulation by U2Af
Nature, 475, 2011
|
|
2ALE
 
 | | Crystal structure of yeast RNA splicing factor Snu13p | | Descriptor: | MAGNESIUM ION, NHP2/L7aE family protein YEL026W | | Authors: | Dobbyn, H.C, McEwan, P.A, Bella, J, O'Keefe, R.T. | | Deposit date: | 2005-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of pre-mRNA and pre-rRNA processing factor Snu13p structure and mutants.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
7LD5
 
 | | polynucleotide phosphorylase | | Descriptor: | MAGNESIUM ION, Polyribonucleotide nucleotidyltransferase, poly-A RNA fragment | | Authors: | Goldgur, Y, Shuman, S, De La Cruz, M.J, Ghosh, S, Unciuleac, M.-C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure and mechanism of Mycobacterium smegmatis polynucleotide phosphorylase.
Rna, 27, 2021
|
|
