3BWE
 
 | | Crystal structure of aggregated form of DJ1 | | Descriptor: | PHOSPHATE ION, Protein DJ-1 | | Authors: | Cha, S.S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
J.Biol.Chem., 283, 2008
|
|
2WTP
 
 | | Crystal Structure of Cu-form Czce from C. metallidurans CH34 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Haertlein, I, Girard, E, Sarret, G, Hazemann, J, Gourhant, P, Kahn, R, Coves, J. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evidence for Conformational Changes Upon Copper Binding to Cupriavidus Metallidurans Czce.
Biochemistry, 49, 2010
|
|
5A8O
 
 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
4HGM
 
 | | Shark IgNAR Variable Domain | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, Serum albumin, ... | | Authors: | Olland, A, Kovalenko, O.V, King, D, Svenson, K. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Atypical Antigen Recognition Mode of a Shark Immunoglobulin New Antigen Receptor (IgNAR) Variable Domain Characterized by Humanization and Structural Analysis.
J.Biol.Chem., 288, 2013
|
|
3EGQ
 
 | |
3WS8
 
 | | Crystal structure of PDE10A in complex with a benzimidazole inhibitor | | Descriptor: | 8-methyl-6-[2-(5-methyl-1-phenyl-1H-benzimidazol-2-yl)ethyl]imidazo[1,5-a]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel benzimidazole derivatives as phosphodiesterase 10A (PDE10A) inhibitors with improved metabolic stability.
Bioorg.Med.Chem., 22, 2014
|
|
1HN0
 
 | |
5ZCD
 
 | | Crystal structure of Alpha-glucosidase in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
1RYA
 
 | | Crystal Structure of the E. coli GDP-mannose mannosyl hydrolase in complex with GDP and MG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GDP-mannose mannosyl hydrolase, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Legler, P.M, Mildvan, A.S, Amzel, L.M. | | Deposit date: | 2003-12-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus.
Structure, 12, 2004
|
|
7JLK
 
 | |
6C5J
 
 | | S25-23 Fab in complex with Chlamydiaceae LPS (Crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, IgG1 Fab Heavy Chain, ... | | Authors: | Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Subtle Changes in the Combining Site of the Chlamydiaceae-Specific mAb S25-23 Increase the Antibody-Carbohydrate Binding Affinity by an Order of Magnitude.
Biochemistry, 58, 2019
|
|
4KDX
 
 | | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group p21, form(1) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase domain | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group P21, form(1)
To be Published
|
|
3B3G
 
 | | The 2.4 A crystal structure of the apo catalytic domain of coactivator-associated arginine methyl transferase I(CARM1,140-480). | | Descriptor: | Histone-arginine methyltransferase CARM1 | | Authors: | Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, Cavarelli, J. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.
Embo J., 26, 2007
|
|
5SV2
 
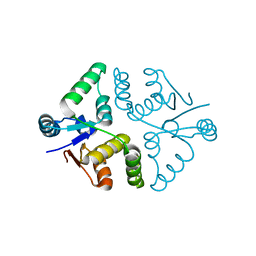 | | Toxin VapC21 from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease VapC21 | | Authors: | Valadares, N.F. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystal structure of VapC21 from Mycobacterium tuberculosis at 1.31 angstrom resolution.
Biochem.Biophys.Res.Commun., 478, 2016
|
|
5A6M
 
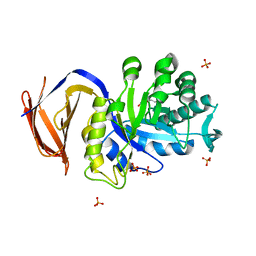 | | Determining the specificities of the catalytic site from the very high resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with a xylotetraose bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4FLO
 
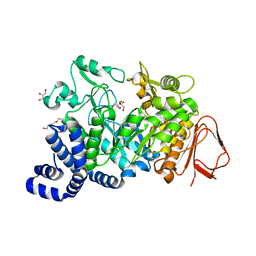 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
7JY0
 
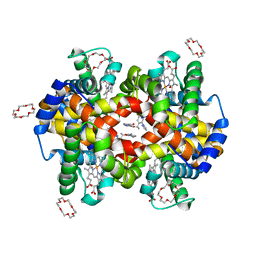 | | Structure of HbA with compound 9 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2-amino-3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}quinoline-6-carboxamide, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
3E7V
 
 | | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-aminophenyl)-N-(3-chlorophenyl)pyrazolo[1,5-a]pyrimidin-5-amine, NICKEL (II) ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand
To be Published
|
|
1HX6
 
 | | P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAJOR CAPSID PROTEIN, ... | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5EJM
 
 | | ThDP-Mn2+ complex of R413A variant of EcMenD soaked with 2-ketoglutarate for 35 min | | Descriptor: | (4~{R})-4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD, a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 2018
|
|
4PZ4
 
 | |
5EJ7
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 21 s | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, GLYCEROL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
3SL8
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4Q1F
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 12R {N-{2-[5-(4-{(1R)-1-[(4,6-diaminopyrimidin-2-yl)sulfanyl]ethyl}-5-methyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]ethyl}methanesulfonamide} | | Descriptor: | Deoxycytidine kinase, N-{2-[5-(4-{(1R)-1-[(4,6-diaminopyrimidin-2-yl)sulfanyl]ethyl}-5-methyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]ethyl}methanesulfonamide, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
2P82
 
 | | Cysteine protease ATG4A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cysteine protease ATG4A | | Authors: | Walker, J.R, Davis, T, Mujib, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human cysteine protease ATG4A
To be Published
|
|
