5LU8
 
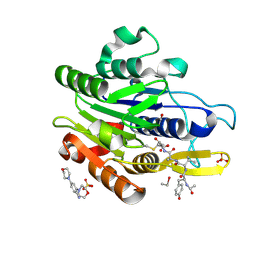 | | CRYSTAL STRUCTURE OF YVAD-CMK BOUND HUMAN LEGUMAIN (AEP) IN COMPLEX WITH COMPOUND 11B | | Descriptor: | 2,4-di(morpholin-4-yl)aniline, 2-acetamido-2-deoxy-beta-D-glucopyranose, AC-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
7REG
 
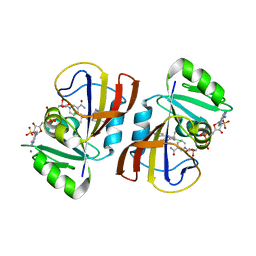 | | DfrA1 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, CALCIUM ION, Dihydrofolate reductase type 1, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
5IO9
 
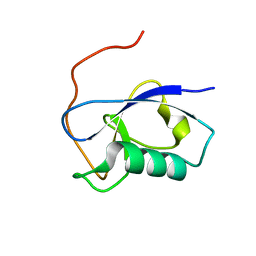 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6DKQ
 
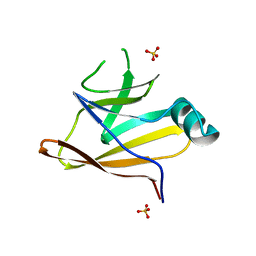 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 | | Descriptor: | Heme-binding protein Shr, SULFATE ION | | Authors: | Macdonald, R, Cascio, D, Collazo, M.J, Clubb, R.T. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Streptococcus pyogenes Shr protein captures human hemoglobin using two structurally unique binding domains.
J.Biol.Chem., 293, 2018
|
|
2ENB
 
 | |
1LI1
 
 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6NGS
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(3-(dimethylamino)propyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[3-(dimethylamino)propyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
6NH4
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(3-fluoro-5-(2-((2R,4S)-4-fluoropyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R,4S)-4-fluoropyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
5LRT
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP and Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Bolten, M, Vahlensieck, C, Lipp, C, Leibundgut, M, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-08-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Depupylase Dop Requires Inorganic Phosphate in the Active Site for Catalysis.
J. Biol. Chem., 292, 2017
|
|
5UL5
 
 | | Crystal structure of RPE65 in complex with MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Tuning of Visual Cycle Modulator Pharmacodynamics.
J. Pharmacol. Exp. Ther., 362, 2017
|
|
6QXZ
 
 | | Solution structure of the ASHH2 CW domain with the N-terminal histone H3 tail mimicking peptide monomethylated on lysine 4 | | Descriptor: | ALA-ARG-THR-MLZ-GLN-THR-ALA-ARG-TYR, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Dobrovolska, O, Madeleine, N, Teigen, K, Halskau, O, Bril'kov, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Febs J., 287, 2020
|
|
5ISX
 
 | |
6B9T
 
 | |
9B3F
 
 | | Cryo-EM structure of yeast (Nap1)2-H2A-H2B-Kap114 | | Descriptor: | Histone H2A, Histone H2B, KAP114 isoform 1, ... | | Authors: | Jiou, J, Fung, H.Y.J, Chook, Y.M. | | Deposit date: | 2024-03-19 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Nap1 and Kap114 co-chaperone H2A-H2B and facilitate targeted histone release in the nucleus.
J.Cell Biol., 224, 2025
|
|
6QXD
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with JKB inhibitor in the active site. | | Descriptor: | (2,4-dinitrophenyl)-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]methanone, COPPER (II) ION, Tyrosinase | | Authors: | Deri Zenaty, B, Gitto, R, Pazy, Y, Fishman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Exploiting the 1-(4-fluorobenzyl)piperazine fragment for the development of novel tyrosinase inhibitors as anti-melanogenic agents: Design, synthesis, structural insights and biological profile.
Eur.J.Med.Chem., 178, 2019
|
|
5EQ3
 
 | | Crystal structure of the SrpA adhesin from Streptococcus sanguinis with a sialyl galactose disaccharide bound | | Descriptor: | ACETATE ION, CALCIUM ION, N-glycolyl-alpha-neuraminic acid-(2-3)-methyl beta-D-galactopyranoside, ... | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5I0F
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with covalent intermediate | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 31, TRIETHYLENE GLYCOL, ... | | Authors: | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5LR3
 
 | |
5I1B
 
 | |
5LR5
 
 | |
5I1Y
 
 | | NvPizza2-H16S58 with cobalt | | Descriptor: | COBALT (II) ION, SULFATE ION, nvPizz2a-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NvPizza2-H16S58 with cobalt
To Be Published
|
|
5LU9
 
 | | Crystal structure of YVAD-cmk bound human legumain (AEP) in complex with compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(morpholin-4-yl)-2,1,3-benzoxadiazol-4-amine, AC-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
5EJ2
 
 | |
7RGS
 
 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
