2OYQ
 
 | | Crystal structure of RB69 gp43 in complex with DNA with 5-NIMP opposite an abasic site analog | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, DNA polymerase, MAGNESIUM ION, ... | | Authors: | Zahn, K.E, Belrhali, H, Wallace, S.S, Doublie, S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Caught bending the a-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide.
Biochemistry, 46, 2007
|
|
4LZB
 
 | | Uracil binding pocket in Vaccinia virus uracil DNA glycosylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the uracil complex of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1UA0
 
 | | Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
3ABE
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
2DTU
 
 | | Crystal structure of the beta hairpin loop deletion variant of RB69 gp43 in complex with DNA containing an abasic site analog | | Descriptor: | 5'-D(*CP*GP*(3DR)P*CP*TP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*AP*GP*A)-3', DNA polymerase | | Authors: | Aller, P, Hogg, M, Konigsberg, W, Wallace, S.S, Doublie, S. | | Deposit date: | 2006-07-15 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family.
J.Biol.Chem., 282, 2007
|
|
7NPF
 
 | | Vibrio cholerae ParA2-ATPyS-DNA filament | | Descriptor: | AAA family ATPase, DNA (49-MER), MAGNESIUM ION, ... | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
5CBX
 
 | | AncGR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2OR1
 
 | | RECOGNITION OF A DNA OPERATOR BY THE REPRESSOR OF PHAGE 434. A VIEW AT HIGH RESOLUTION | | Descriptor: | 434 REPRESSOR, DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3') | | Authors: | Aggarwal, A.K, Rodgers, D.W, Drottar, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1989-09-05 | | Release date: | 1989-09-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a DNA operator by the repressor of phage 434: a view at high resolution.
Science, 242, 1988
|
|
6Z1A
 
 | | Ternary complex of Staphylococcus aureus DNA gyrase with AMK12 and DNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kolaric, A, Germe, T, Hrast, M, Stevenson, C.E.M, Lawson, D.M, Burton, N, Voros, J, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds.
Nat Commun, 12, 2021
|
|
1D37
 
 | |
7NX5
 
 | |
8QAM
 
 | |
2D5V
 
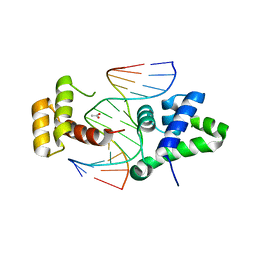 | | Crystal structure of HNF-6alpha DNA-binding domain in complex with the TTR promoter | | Descriptor: | 5'-D(*AP*TP*TP*AP*TP*TP*GP*AP*CP*TP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*AP*GP*TP*CP*AP*AP*TP*AP*AP*T)-3', ACETATE ION, ... | | Authors: | Iyaguchi, D, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2005-11-07 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6
Structure, 15, 2007
|
|
5OS9
 
 | |
362D
 
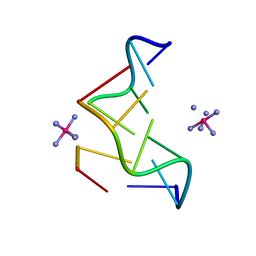 | | THE STRUCTURE OF D(TGCGCA)2 AND A COMPARISON TO OTHER Z-DNA HEXAMERS | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*TP*GP*CP*GP*CP*A)-3') | | Authors: | Harper, N.A, Brannigan, J.A, Buck, M, Lewis, R.J, Moore, M.H, Schneider, B. | | Deposit date: | 1997-08-20 | | Release date: | 1997-11-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of d(TGCGCA)2 and a comparison to other DNA hexamers.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2DES
 
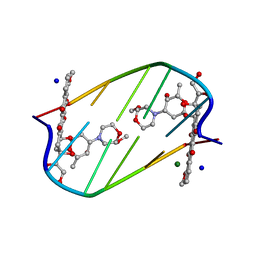 | | INTERACTIONS BETWEEN MORPHOLINYL ANTHRACYCLINES AND DNA: THE CRYSTAL STRUCTURE OF A MORPHOLINO DOXORUBICIN BOUND TO D(CGTACG) | | Descriptor: | 3'-DESAMINO-3'-(2-METHOXY-4-MORPHOLINYL)-DOXORUBICIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Cirilli, M, Bachechi, F, Ughetto, G, Colonna, F.P, Capobianco, M.L. | | Deposit date: | 1993-03-16 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions between morpholinyl anthracyclines and DNA. The crystal structure of a morpholino doxorubicin bound to d(CGTACG).
J.Mol.Biol., 230, 1993
|
|
5BOX
 
 | | Structure of TrmBL2, an archaeal chromatin protein, shows a novel mode of DNA binding. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (25-MER), DNA TGM (25-MER), ... | | Authors: | Ahmad, M.U, Diederichs, K, Welte, W. | | Deposit date: | 2015-05-27 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Nonspecific Binding of DNA by TrmBL2, an Archaeal Chromatin Protein.
J.Mol.Biol., 427, 2015
|
|
6QX1
 
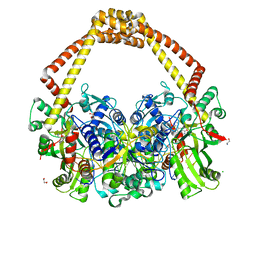 | | 2.7A structure of benzoisoxazole 3 with S.aureus DNA gyrase and DNA. | | Descriptor: | (2~{R})-2-[[5-(2-chlorophenyl)-1,2-benzoxazol-3-yl]oxy]-2-phenyl-ethanamine, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-03-06 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-guided design of antibacterials that allosterically inhibit DNA gyrase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6L84
 
 | | Complex of DNA polymerase IV and D-DNA duplex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(P*CP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
5B43
 
 | | Crystal structure of Acidaminococcus sp. Cpf1 in complex with crRNA and target DNA | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cpf1, DNA (34-MER), ... | | Authors: | Yamano, T, Nishimasu, H, Hirano, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA
Cell, 165, 2016
|
|
5CA7
 
 | |
4DQY
 
 | |
3NGD
 
 | |
4WS6
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-aminouracil, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WPL
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form I | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
