8U5Y
 
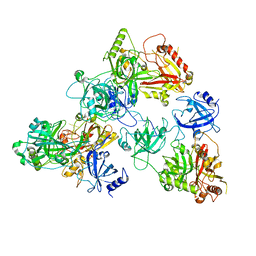 | | human RADX trimer bound to ssDNA | | Descriptor: | DNA (25-MER), RPA-related protein RADX | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Biorxiv, 2023
|
|
8U5T
 
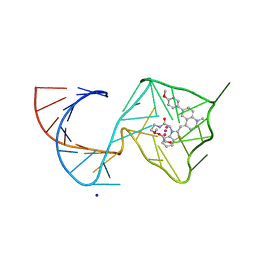 | | Structure of Mango II variant aptamer bound to T01-6A-B | | Descriptor: | 3-{2,16-dioxo-20-[(3aS,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-6A-B
To Be Published
|
|
8U5R
 
 | |
8U5P
 
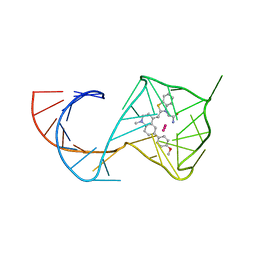 | | Structure of Mango II aptamer bound to T01-6A-B | | Descriptor: | 3-[2,16-dioxo-20-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-6,9,12-trioxa-3,15-diazaicosan-1-yl]-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Mango II aptamer bound to T01-6A-B
To Be Published
|
|
8U5M
 
 | | Structure of Sts-1 HP domain with rebamipide | | Descriptor: | Rebamipide, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Azia, F, Dey, R, French, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
8U5K
 
 | |
8U5J
 
 | | Structure of Mango III variant aptamer bound to T01-07M-B | | Descriptor: | 2-[(E)-(1,7-dimethylquinolin-4(1H)-ylidene)methyl]-3-{2,16-dioxo-20-[(3aR,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium, DIMETHYL SULFOXIDE, Mango III variant, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mango III variant aptamer bound to T01-07M-B
To Be Published
|
|
8U5H
 
 | |
8U5G
 
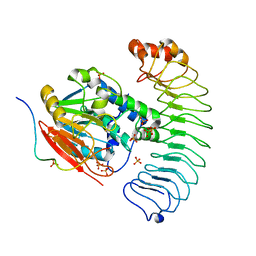 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
8U5F
 
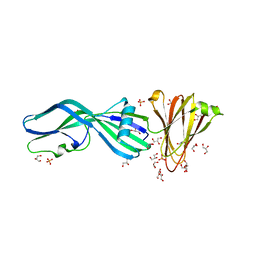 | | Crystal Structure of Trypsinized Clostridium perfringens Enterotoxin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Kapoor, S, Ogbu, C.P, Vecchio, A.J. | | Deposit date: | 2023-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin.
Toxins, 15, 2023
|
|
8U5E
 
 | |
8U5D
 
 | |
8U5B
 
 | |
8U5A
 
 | | Improving protein expression, stability, and function with ProteinMPNN | | Descriptor: | Designed myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kalvet, I, Bera, A.K, Baker, D. | | Deposit date: | 2023-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving Protein Expression, Stability, and Function with ProteinMPNN.
J.Am.Chem.Soc., 146, 2024
|
|
8U55
 
 | |
8U53
 
 | | Mechanically activated ion channel OSCA3.1 in nanodiscs | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CSC1-like protein ERD4, PALMITIC ACID, ... | | Authors: | Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli.
Elife, 12, 2024
|
|
8U51
 
 | |
8U50
 
 | |
8U4Z
 
 | |
8U4W
 
 | |
8U4V
 
 | |
8U4U
 
 | |
8U4T
 
 | |
8U4S
 
 | |
8U4R
 
 | |
