5HL8
 
 | | 1.93 Angstrom resolution crystal structure of a pullulanase-specific type II secretion system integral cytoplasmic membrane protein GspL (C-terminal fragment; residues 309-397) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | Type II secretion system protein L | | Authors: | Halavaty, A.S, Minasov, G, Kiryukhina, O, Grimshaw, S, Light, S, Dubrovska, I, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom resolution crystal structure of a pullulanase-specific type II secretion system integral cytoplasmic membrane protein GspL (C-terminal fragment; residues 309-397) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044
To Be Published
|
|
1SPE
 
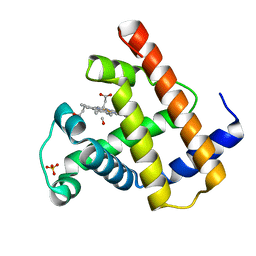 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
6E6P
 
 | | HRAS G13D bound to GppNHp (Ha,b,c13GNP) | | Descriptor: | CALCIUM ION, GLYCEROL, GTPase HRas, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
5MBA
 
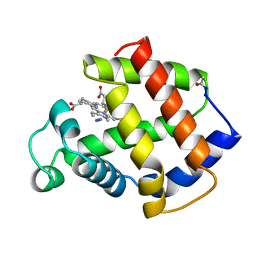 | | BINDING MODE OF AZIDE TO FERRIC APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | AZIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1991-01-14 | | Release date: | 1992-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding mode of azide to ferric Aplysia limacina myoglobin. Crystallographic analysis at 1.9 A resolution.
J.Mol.Recog., 4, 1991
|
|
6E6C
 
 | | HRAS G13D bound to GppNHp (H13GNP) | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6E6H
 
 | | NRAS G13D bound to GppNHp (N13GNP) | | Descriptor: | GTPase NRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
1IWH
 
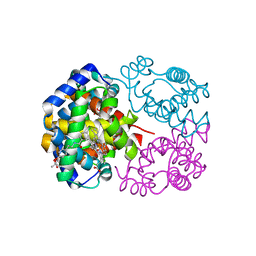 | | Crystal Structure of Horse Carbonmonoxyhemoglobin-Bezafibrate Complex at 1.55A Resolution: A Novel Allosteric Binding Site in R-State Hemoglobin | | Descriptor: | 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, CARBON MONOXIDE, Hemoglobin alpha chain, ... | | Authors: | Shibayama, N, Miura, S, Tame, J.R.H, Yonetani, T, Park, S.-Y. | | Deposit date: | 2002-05-15 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Horse Carbonmonoxyhemoglobin-Bezafibrate Complex at 1.55A Resolution. A Novel Allosteric Binding Site in R-State Hemoglobin
J.BIOL.CHEM., 277, 2002
|
|
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6YXX
 
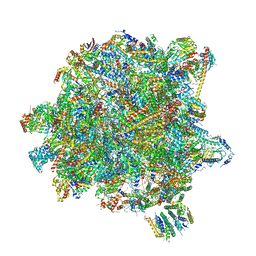 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6Z3T
 
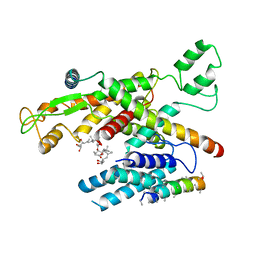 | | Structure of canine Sec61 inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Gerard, S.F, Higgins, M.K. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Inhibited State of the Sec Translocon.
Mol.Cell, 79, 2020
|
|
5O8O
 
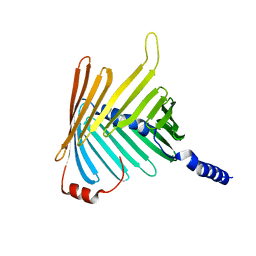 | | N. crassa Tom40 model based on cryo-EM structure of the TOM core complex at 6.8 A | | Descriptor: | Mitochondrial import receptor subunit tom40 | | Authors: | Bausewein, T, Mills, D.J, Nussberger, S, Nitschke, B, Kuehlbrandt, W. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM Structure of the TOM Core Complex from Neurospora crassa.
Cell, 170, 2017
|
|
6F0E
 
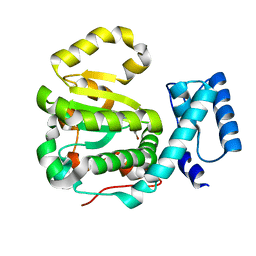 | | Structure of yeast Sec14p with a picolinamide compound | | Descriptor: | SEC14 cytosolic factor, ~{N}-(1,3-benzodioxol-5-ylmethyl)-5-bromanyl-3-fluoranyl-pyridine-2-carboxamide | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties.
Cell Chem Biol, 25, 2018
|
|
5OE7
 
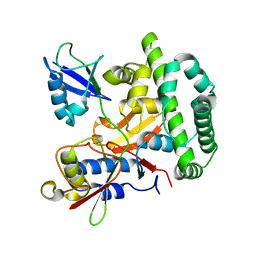 | |
6FKR
 
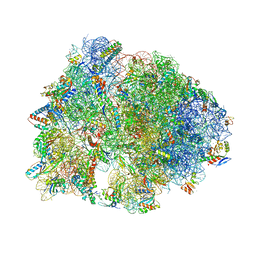 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
7AE9
 
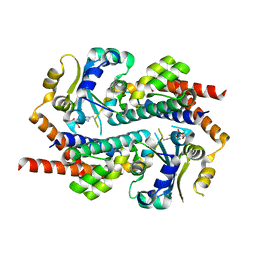 | | Crystal structure of mono-AMPylated HEPN(R46E) toxin in complex with MNT antitoxin | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-MONOPHOSPHATE, HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE8
 
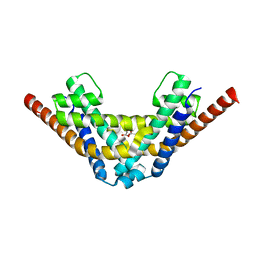 | | Crystal structure of HEPN(R102A) toxin | | Descriptor: | ACETATE ION, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AER
 
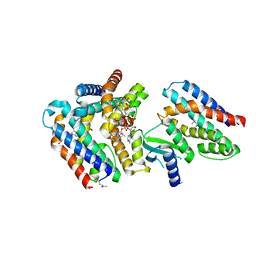 | | Rebuilt and re-refined PDB entry 5yep: tri-AMPylated Shewanella oneidensis HEPN toxin in complex with MNT antitoxin | | Descriptor: | ADENOSINE MONOPHOSPHATE, Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
5OA9
 
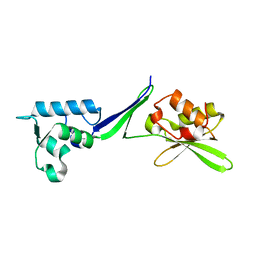 | | Human translation re-initiation complex containing eIF2D | | Descriptor: | Eukaryotic translation initiation factor 2D | | Authors: | Weisser, M, Schaefer, T, Leibundgut, M, Boehringer, D, Aylett, C.H.S, Ban, N. | | Deposit date: | 2017-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insights into Human Re-initiation Complexes.
Mol. Cell, 67, 2017
|
|
5OA3
 
 | | Human 40S-eIF2D-re-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Weisser, M, Schaefer, T, Leibundgut, M, Boehringer, D, Aylett, C.H.S, Ban, N. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and Functional Insights into Human Re-initiation Complexes.
Mol. Cell, 67, 2017
|
|
7AE2
 
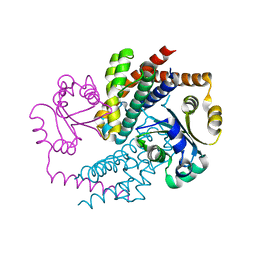 | | Crystal structure of HEPN(H107A-Y109F) toxin in complex with MNT antitoxin | | Descriptor: | HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE6
 
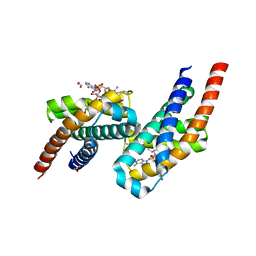 | | Crystal structure of di-AMPylated HEPN(R102A) toxin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
5OAP
 
 | |
5OAO
 
 | |
6E6F
 
 | | KRAS G13D bound to GppNHp (K13GNP) | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6E6G
 
 | | KRAS G13D bound to GDP (K13GDP) | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
