6K8P
 
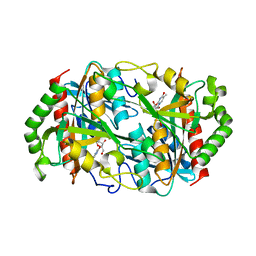 | |
1OBW
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Oganessyan, V.Yu, Harutyunyan, E.H, Avaeva, S.M, Oganessyan, N.N, Mather, T, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Biochemistry, 36, 1997
|
|
6KEH
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 6,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEC
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEJ
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6-[2-(diethylamino)ethoxy]-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 6-[2-(diethylamino)ethoxy]-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
3LD3
 
 | |
2VWX
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-chloro-1,3-benzodioxol-4-yl)amino]pyrimidin-2-yl}amino)benzenesulfonamide, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4P9T
 
 | | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin | | Descriptor: | 1,2-ETHANEDIOL, Catenin alpha-2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shibahara, T, Hirano, Y, Hakoshima, T. | | Deposit date: | 2014-04-04 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin.
Febs Lett., 589, 2015
|
|
4ZS9
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sugar binding protein of ABC transporter system, ... | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
7GBA
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ANT-OPE-d972fbad-1 (Mpro-x10296) | | Descriptor: | 1-{4-[(4-fluorophenyl)methyl]piperazin-1-yl}propan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5XOS
 
 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
1ZAR
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ADP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
2VWZ
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-[3-[[4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL]AMINO]PHENYL]METHANESULFONAMIDE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VX1
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL}AMINO)BENZAMIDE, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
6IYG
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
4CD3
 
 | | RnNTPDase2 X4 variant in complex with PSB-071 | | Descriptor: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
4PP5
 
 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 5-methyl UAB30 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-(5-methyl-3,4-dihydronaphthalen-1(2H)-ylidene)octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity.
J.Med.Chem., 57, 2014
|
|
2OIY
 
 | |
1LQO
 
 | | Crystal Strutcure of the Fosfomycin Resistance Protein A (FosA) Containing Bound Thallium Cations | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROBABLE Fosfomycin Resistance Protein, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
3S4C
 
 | | Lactose phosphorylase in complex with sulfate | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Lactose Phosphorylase, SULFATE ION | | Authors: | Van Hoorebeke, A, Stout, J, Soetaert, W, Van Beeumen, J, Desmet, T, Savvides, S. | | Deposit date: | 2011-05-19 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellobiose phosphorylase: reconstructing the structural itinerary along the catalytic pathway
To be Published
|
|
4Z4Y
 
 | | Crystal structure of GII.10 P domain in complex with 7.5mM B antigen (trisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4C1E
 
 | | Crystal structure of the metallo-beta-lactamase VIM-2 with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE CLASS B VIM-2, FORMIC ACID, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
1P7P
 
 | | Methionyl-tRNA synthetase from Escherichia coli complexed with methionine phosphonate | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHONIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-05 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1LCX
 
 | | NMR structure of HIV-1 gp41 659-671 13mer peptide | | Descriptor: | GP41 | | Authors: | Biron, Z, Khare, S, Samson, A.O, Hayek, Y, Naider, F, Anglister, J. | | Deposit date: | 2002-04-07 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Monomeric 3(10)-Helix Is Formed in Water by a 13-Residue Peptide
Representing the Neutralizing Determinant of HIV-1 on gp41.
Biochemistry, 41, 2002
|
|
