5M7G
 
 | | Tubulin-MTD147 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(2,6-dimorpholin-4-ylpyridin-4-yl)-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5M7E
 
 | | Tubulin-BKM120 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5PWC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 in complex with E48115b | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dichloro-N-(pyridin-3-yl)benzamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5FZ1
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 2,4-dichloro-N-pyridin-3-ylbenzamide (E48115b) (ligand modelled based on PANDDA event map) | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dichloro-N-(pyridin-3-yl)benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with E48115B
To be Published
|
|
5FZ4
 
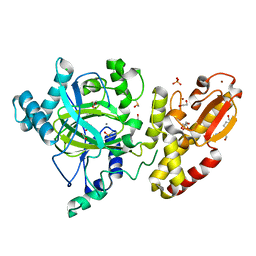 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment (3R)-1-[(3-phenyl-1,2,4-oxadiazol-5-yl)methyl]pyrrolidin-3-ol (N10057a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | (3S)-1-[(3-phenyl-1,2,4-oxadiazol-5-yl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Pearce, N, Talon, R, Collins, P, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N10057A
To be Published
|
|
8DIQ
 
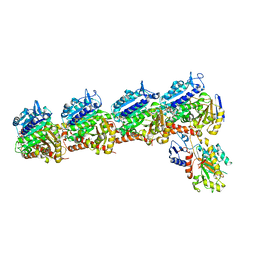 | | Tubulin-RB3_SLD-TTL in complex with SB226 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SB226, an inhibitor of tubulin polymerization, inhibits paclitaxel-resistant melanoma growth and spontaneous metastasis.
Cancer Lett., 555, 2022
|
|
6X1C
 
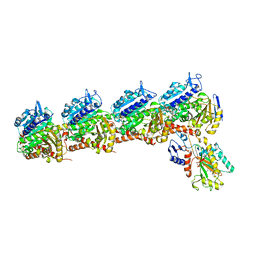 | | Tubulin-RB3_SLD-TTL in complex with compound 5j | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1F
 
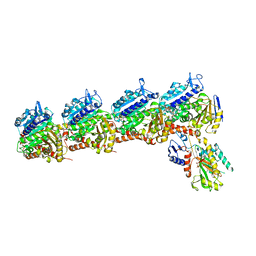 | | Tubulin-RB3_SLD-TTL in complex with compound 5m | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methoxy-4-(2-methyl-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1E
 
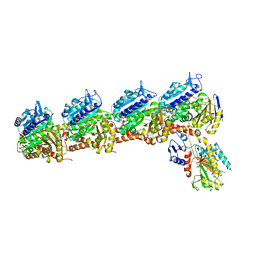 | | Tubulin-RB3_SLD-TTL in complex with compound 5l | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloro-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
3H2H
 
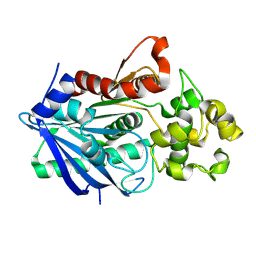 | |
6KM1
 
 | | Human Carbonic Anhydrase II V143I variant 13 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM4
 
 | | Human Carbonic Anhydrase II native 07 atm CO2 | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
3H53
 
 | | Crystal Structure of human alpha-N-acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminidase, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2009-04-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases
J.Mol.Biol., 393, 2009
|
|
4QSB
 
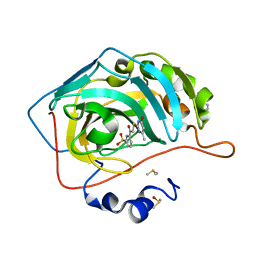 | | Crystal structure of human carbonic anhydrase isozyme II with 3-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)thio]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-07-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Intrinsic Thermodynamics and Structure Correlation of Benzenesulfonamides with a Pyrimidine Moiety Binding to Carbonic Anhydrases I, II, VII, XII, and XIII
Plos One, 9, 2014
|
|
3H2I
 
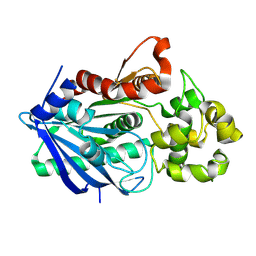 | |
3X41
 
 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
1GY9
 
 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Burzlaff, N.I, Ryle, M.J, Lloyd, J.S, Clifton, I.J, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2002-04-22 | | Release date: | 2002-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates.
Biochemistry, 41, 2002
|
|
4OQE
 
 | | Crystal structure of the tylM1 N,N-dimethyltransferase in complex with SAH and TDP-Fuc3NMe | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, dTDP-3-N-methylamino-3,6-dideoxygalactose, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Production of a novel N-monomethylated dideoxysugar.
Biochemistry, 53, 2014
|
|
4IOM
 
 | | N10-formyltetrahydrofolate synthetase from Moorella thermoacetica with folate | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, FOLIC ACID, Formate--tetrahydrofolate ligase, ... | | Authors: | Stec, B. | | Deposit date: | 2013-01-08 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Time passes yet errors remain: Comments on the structure of N(10) -formyltetrahydrofolate synthetase.
Protein Sci., 22, 2013
|
|
6KLZ
 
 | | Human Carbonic Anhydrase II V143I variant 00 atm CO2 | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM3
 
 | | Human Carbonic Anhydrase II native 00 atm CO2 | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
6KM5
 
 | | Human Carbonic Anhydrase II native 13 atm CO2 | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
3H2G
 
 | |
5M8G
 
 | | Tubulin-MTD265 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(2-morpholin-4-yl-6-pyrrolidin-1-yl-pyrimidin-4-yl)-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
6KM0
 
 | | Human Carbonic Anhydrase II V143I variant 07 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
