8W79
 
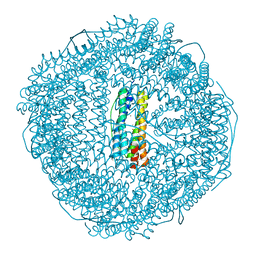 | | Fe-O nanocluster of form-III in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
5AC8
 
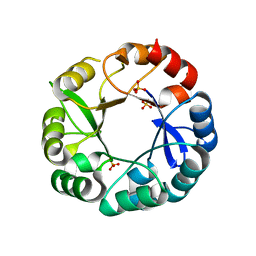 | | S. enterica HisA with mutations D10G, dup13-15, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8W7B
 
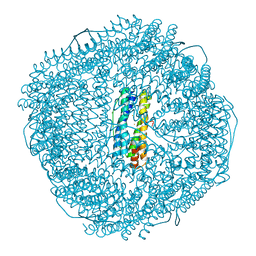 | | Fe-O nanocluster of form-IV in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7T
 
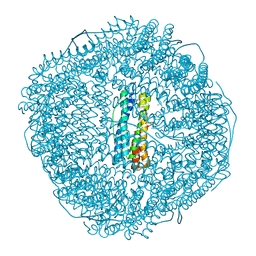 | | Fe-O nanocluster of form-VII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7U
 
 | | Mutant of ferritin from Ureaplasma diversum (Udif-E164A-E168A) without soaking | | Descriptor: | FE (III) ION, Ferritin, MAGNESIUM ION | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7V
 
 | | Udif-E164A-E168A soaking in Fe2+ solution for 50 minutes | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
3ZZ5
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 74 | | Descriptor: | 3C PROTEINASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
8WPT
 
 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum | | Descriptor: | CHLORIDE ION, FE (III) ION, Truncated ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
3ZZ3
 
 | | Crystal structure of 3C protease mutant (N126Y) of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
5U1R
 
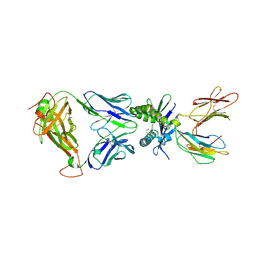 | |
8WPV
 
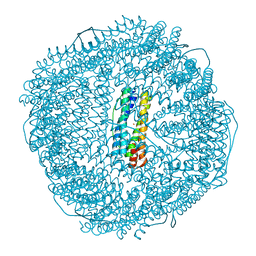 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum soaked in Fe2+ solution for 30min | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
3ZZB
 
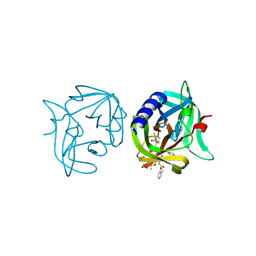 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 85 | | Descriptor: | 3C PROTEINASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
8W7Q
 
 | | Fe-O nanocluster of form-VI in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
3ZPS
 
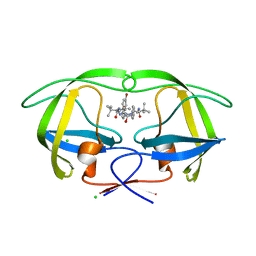 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[(4R)-5-[[(2S)-3,3-dimethyl-1-oxidanylidene-1-(prop-2-enylamino)butan-2-yl]amino]-4-oxidanyl-5-oxidanylidene-4-(phenylmethyl)pentyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors.
J.Med.Chem., 56, 2013
|
|
3ZPU
 
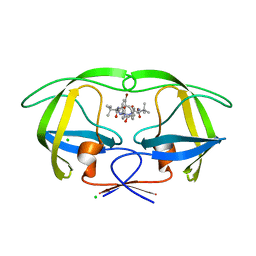 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[3-[(3Z,8S,11R)-8-tert-butyl-11-oxidanyl-7,10-bis(oxidanylidene)-6,9-diazabicyclo[11.2.2]heptadeca-1(15),3,13,16-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | Deposit date: | 2013-03-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors
J.Med.Chem., 56, 2013
|
|
3ZZD
 
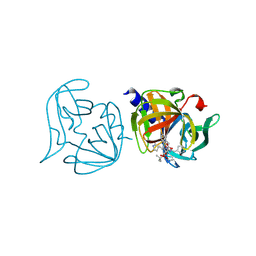 | | Crystal structure of 3C protease mutant (T68A and N126Y) of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 85 | | Descriptor: | 3C PROTEINASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
5U4P
 
 | | Protein-protein complex between 26S proteasome regulatory subunit RPN8, RPN11, and Ubiquitin S31 | | Descriptor: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8, Ubiquitin-40S ribosomal protein S31, ... | | Authors: | Worden, E.J, Dong, K.C, Martin, A. | | Deposit date: | 2016-12-05 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome.
Mol. Cell, 67, 2017
|
|
5FMZ
 
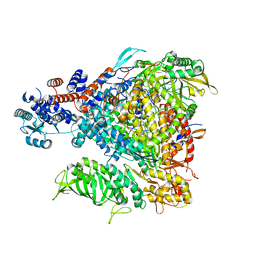 | | Crystal structure of Influenza B polymerase with bound 5' vRNA | | Descriptor: | 5'-R(*AP*GP*UP*AP*GP*UP*AP*AP*CP*AP*AP*GP)-3', POLYMERASE ACIDIC PROTEIN, POLYMERASE BASIC PROTEIN 2, ... | | Authors: | Guilligay, D, Cusack, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
2E02
 
 | |
3ZZA
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 84 | | Descriptor: | 3C PROTEINASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
8WMV
 
 | | The structure of PSI-14CAC complex at stationary growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
3ZZ7
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 81 | | Descriptor: | 3C PROTEINASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
1Z1V
 
 | | NMR structure of the Saccharomyces cerevisiae Ste50 SAM domain | | Descriptor: | STE50 protein | | Authors: | Kwan, J.J, Warner, N, Maini, J, Pawson, T, Donaldson, L.W. | | Deposit date: | 2005-03-06 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Saccharomyces cerevisiae Ste50 binds the MAPKKK Ste11 through a head-to-tail SAM domain interaction.
J.Mol.Biol., 356, 2006
|
|
3ZZ6
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with Michael receptor inhibitor 75 | | Descriptor: | ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE, POLYPROTEIN 3BCD | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Peptidic Ab-Nonsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ4
 
 | | Crystal structure of 3C protease mutant (T68A and N126Y) of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
