1YNB
 
 | | crystal structure of genomics APC5600 | | Descriptor: | hypothetical protein AF1432 | | Authors: | Dong, A, Skarina, T, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of genomics AF1432 by Sulfur SAD methods
To be Published
|
|
1YNC
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YND
 
 | | Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6A resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, SANGLIFEHRIN A | | Authors: | Kallen, J, Sedrani, R, Zenke, G, Wagner, J. | | Deposit date: | 2005-01-24 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6 A resolution.
J.Biol.Chem., 280, 2005
|
|
1YNE
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | APOLIPOPROTEIN B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNH
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNJ
 
 | | Taq RNA polymerase-Sorangicin complex | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Campbell, E.A, Pavlova, O, Zenkin, N, Leon, F, Irschik, H, Jansen, R, Severinov, K, Darst, S.A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase
Embo J., 24, 2005
|
|
1YNK
 
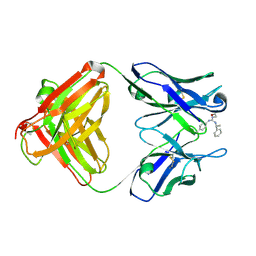 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeteners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-[((R)-{[4-(AMINOMETHYL)PHENYL]AMINO}{[(1R)-1-PHENYLETHYL]AMINO}METHYL)AMINO]ETHANE-1,1-DIOL, Ig gamma heavy chain, immunoglobulin kappa light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
1YNL
 
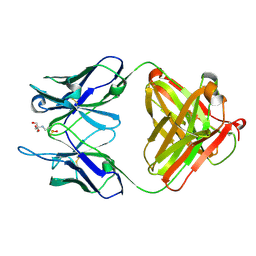 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeterners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Ig gamma heavy chain, Ig gamma light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
1YNM
 
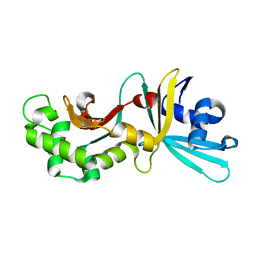 | | Crystal structure of restriction endonuclease HinP1I | | Descriptor: | R.HinP1I restriction endonuclease | | Authors: | Yang, Z, Horton, J.R, Maunus, R, Wilson, G.G, Roberts, R.J, Cheng, X. | | Deposit date: | 2005-01-24 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI
Nucleic Acids Res., 33, 2005
|
|
1YNN
 
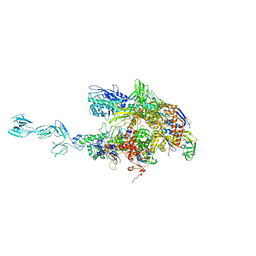 | | Taq RNA polymerase-rifampicin complex | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Campbell, E.A, Pavlova, O, Zenkin, N, Leon, F, Irschik, H, Jansen, R, Severinov, K, Darst, S.A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase
Embo J., 24, 2005
|
|
1YNO
 
 | |
1YNP
 
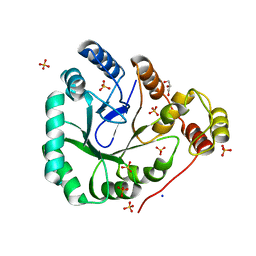 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (apo form) | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
1YNQ
 
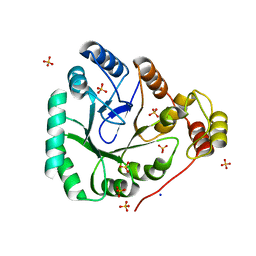 | | aldo-keto reductase AKR11C1 from Bacillus halodurans (holo form) | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Marquardt, T, Kostrewa, D, Winkler, F.K, Li, X.D. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution Crystal Structure of AKR11C1 from Bacillus halodurans: An NADPH-dependent 4-Hydroxy-2,3-trans-nonenal Reductase
J.Mol.Biol., 354, 2005
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
1YNS
 
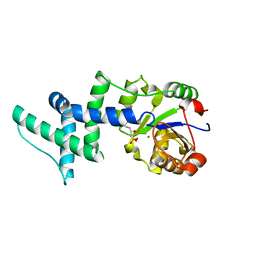 | | Crystal Structure Of Human Enolase-phosphatase E1 and its complex with a substrate analog | | Descriptor: | 2-OXOHEPTYLPHOSPHONIC ACID, E-1 enzyme, MAGNESIUM ION | | Authors: | Wang, H, Pang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human e1 enzyme and its complex with a substrate analog reveals the mechanism of its phosphatase/enolase
J.Mol.Biol., 348, 2005
|
|
1YNT
 
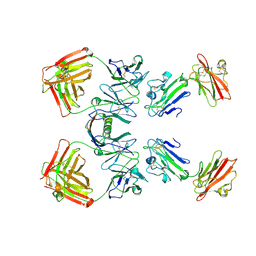 | | Structure of the monomeric form of T. gondii SAG1 surface antigen bound to a human Fab | | Descriptor: | 4F11E12 Fab variable heavy chain region, 4F11E12 Fab variable light chain region, CADMIUM ION, ... | | Authors: | Graille, M, Stura, E.A, Bossus, M, Muller, B.H, Letourneur, O, Battail-Poirot, N, Sibai, G, Rolland, D, Le Du, M.H, Ducancel, F. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex between the monomeric form of Toxoplasma gondii surface antigen 1 (SAG1) and a monoclonal antibody that mimics the human immune response
J.Mol.Biol., 354, 2005
|
|
1YNU
 
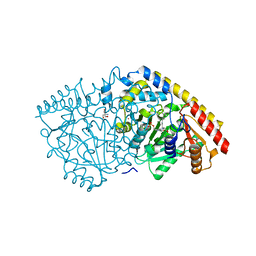 | | Crystal structure of apple ACC synthase in complex with L-vinylglycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ... | | Authors: | Capitani, G, Tschopp, M, Eliot, A.C, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of ACC synthase inactivated by the mechanism-based inhibitor L-vinylglycine.
Febs Lett., 579, 2005
|
|
1YNV
 
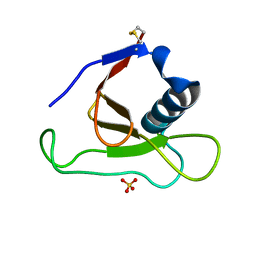 | | Asp79 makes a large, unfavorable contribution to the stability of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Trevino, S.R, Gokulan, K, Newsom, S, Thurlkill, R.L, Shaw, K.L, Mitkevich, V.A, Makarov, A.A, Sacchettini, J.C, Scholtz, J.M, Pace, C.N. | | Deposit date: | 2005-01-25 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Asp79 Makes a Large, Unfavorable Contribution to the Stability of RNase Sa.
J.Mol.Biol., 354, 2005
|
|
1YNW
 
 | |
1YNX
 
 | |
1YNY
 
 | | Molecular Structure of D-Hydantoinase from a Bacillus sp. AR9: Evidence for mercury inhibition | | Descriptor: | D-Hydantoinase, MANGANESE (II) ION | | Authors: | Radha Kishan, K.V, Vohra, R.M, Ganeshan, K, Agrawal, V, Sharma, V.M, Sharma, R. | | Deposit date: | 2005-01-26 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of D-hydantoinase from Bacillus sp. AR9: evidence for mercury inhibition.
J.Mol.Biol., 347, 2005
|
|
1YNZ
 
 | | SH3 domain of yeast Pin3 | | Descriptor: | Pin3p | | Authors: | Kursula, P, Kursula, I, Zou, P, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-01-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the yeast SH3 domain proteome
To be Published
|
|
