1CGE
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, ZINC ION | | Authors: | Lovejoy, B, Hassell, A.M, Luther, M.A, Weigl, D, Jordan, S.R. | | Deposit date: | 1994-02-03 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself.
Biochemistry, 33, 1994
|
|
1MSG
 
 | |
194D
 
 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3') | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1CGF
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, ZINC ION | | Authors: | Lovejoy, B, Hassell, A.M, Luther, M.A, Weigl, D, Jordan, S.R. | | Deposit date: | 1994-02-03 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself.
Biochemistry, 33, 1994
|
|
2ADA
 
 | |
2AYH
 
 | | CRYSTAL AND MOLECULAR STRUCTURE AT 1.6 ANGSTROMS RESOLUTION OF THE HYBRID BACILLUS ENDO-1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE H(A16-M) | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Keitel, T, Heinemann, U. | | Deposit date: | 1995-02-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure at 0.16-nm resolution of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase H(A16-M).
Eur.J.Biochem., 232, 1995
|
|
1OLH
 
 | |
1IVF
 
 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
1FPT
 
 | |
1LMK
 
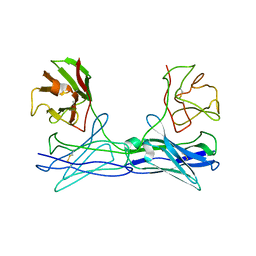 | | THE STRUCTURE OF A BIVALENT DIABODY | | Descriptor: | ANTI-PHOSPHATIDYLINOSITOL SPECIFIC PHOSPHOLIPASE C DIABODY | | Authors: | Williams, R.L. | | Deposit date: | 1994-08-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a diabody, a bivalent antibody fragment.
Structure, 2, 1994
|
|
1LKI
 
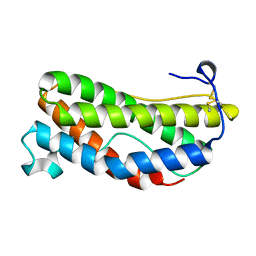 | | THE CRYSTAL STRUCTURE AND BIOLOGICAL FUNCTION OF LEUKEMIA INHIBITORY FACTOR: IMPLICATIONS FOR RECEPTOR BINDING | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Grey, L.M, Staunton, D, Stuart, D.I, Heath, J.K, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding.
Cell(Cambridge,Mass.), 77, 1994
|
|
1PFL
 
 | | REFINED SOLUTION STRUCTURE OF HUMAN PROFILIN I | | Descriptor: | PROFILIN I | | Authors: | Metzler, W.J, Farmer II, B.T, Constantine, K.L, Friedrichs, M.S, Lavoie, T, Mueller, L. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of human profilin I.
Protein Sci., 4, 1995
|
|
1PDH
 
 | |
1HTP
 
 | |
1HPN
 
 | | N.M.R. AND MOLECULAR-MODELLING STUDIES OF THE SOLUTION CONFORMATION OF HEPARIN | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Mulloy, B, Forster, M.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | N.m.r. and molecular-modelling studies of the solution conformation of heparin.
Biochem.J., 293, 1993
|
|
1KAL
 
 | |
1KNB
 
 | |
1IVB
 
 | |
1TMC
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX MOLECULE MISSING THE ALPHA3 DOMAIN OF THE HEAVY CHAIN | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-AW68), DECAMERIC PEPTIDE (EVAPPEYHRK) | | Authors: | Collins, E.J, Garboczi, D.N, Karpusas, M.N, Wiley, D.C. | | Deposit date: | 1994-12-19 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of a class I major histocompatibility complex molecule missing the alpha 3 domain of the heavy chain.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1MSF
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
1MSH
 
 | |
1NHK
 
 | |
1PRH
 
 | |
1PVU
 
 | |
1GFI
 
 | | STRUCTURES OF ACTIVE CONFORMATIONS OF GI ALPHA 1 AND THE MECHANISM OF GTP HYDROLYSIS | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Berghuis, A.M, Sprang, S.R. | | Deposit date: | 1994-11-11 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis.
Science, 265, 1994
|
|
