3EFX
 
 | | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin, 1.9A crystal structure reveals the details | | Descriptor: | Cholera enterotoxin subunit B, Heat-labile enterotoxin B chain, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose | | Authors: | Holmner, A, Lebens, M, Teneberg, S, Angstrom, J, Okvist, M, Krengel, U. | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin: 1.9 A crystal structure reveals the details
Structure, 12, 2004
|
|
4BNA
 
 | |
190L
 
 | |
1AFF
 
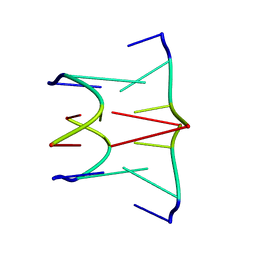 | | DNA QUADRUPLEX CONTAINING GGGG TETRADS AND (T.A).A TRIADS, NMR, 8 STRUCTURES | | Descriptor: | QUADRUPLEX DNA (5'-D(TP*AP*GP*G)-3') | | Authors: | Kettani, A, Bouaziz, S, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads.
Nat.Struct.Biol., 4, 1997
|
|
4KBL
 
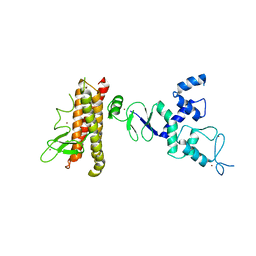 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
1IXI
 
 | |
1IXG
 
 | |
1IXH
 
 | |
1BDM
 
 | |
1SFU
 
 | | Crystal structure of the viral Zalpha domain bound to left-handed Z-DNA | | Descriptor: | 34L protein, 5'-D(*T*CP*GP*CP*GP*CP*G)-3' | | Authors: | Ha, S.C, Van Quyen, D, Wu, C.A, Lowenhaupt, K, Rich, A, Kim, Y.G, Kim, K.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
441D
 
 | |
1BRN
 
 | |
1KXM
 
 | | Crystal structure of Cytochrome c Peroxidase with a Proposed Electron Transfer Pathway Excised to Form a Ligand Binding Channel. | | Descriptor: | BENZIMIDAZOLE, Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rosenfeld, R.J, Hayes, A.M.A, Musah, R.A, Goodin, D.B. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Excision of a proposed electron transfer pathway in cytochrome c peroxidase and its replacement by a ligand-binding channel.
Protein Sci., 11, 2002
|
|
3F8G
 
 | | The X-ray structure of a dimeric variant of human pancreatic ribonuclease with high cytotoxic and antitumor activities | | Descriptor: | Ribonuclease pancreatic, SULFATE ION | | Authors: | Merlino, A, Avella, G, Mazzarella, L, Sica, F. | | Deposit date: | 2008-11-12 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features for the mechanism of antitumor action of a dimeric human pancreatic ribonuclease variant.
Protein Sci., 18, 2009
|
|
1PUT
 
 | | AN NMR-DERIVED MODEL FOR THE SOLUTION STRUCTURE OF OXIDIZED PUTIDAREDOXIN, A 2FE, 2-S FERREDOXIN FROM PSEUDOMONAS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PUTIDAREDOXIN | | Authors: | Pochapsky, T.C, Ye, X.M, Ratnaswamy, G, Lyons, T.A. | | Deposit date: | 1994-07-09 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized putidaredoxin, a 2-Fe, 2-S ferredoxin from Pseudomonas.
Biochemistry, 33, 1994
|
|
2XNY
 
 | | A fragment of streptococcal M1 protein in complex with human fibrinogen | | Descriptor: | FIBRINOGEN ALPHA CHAIN, FIBRINOGEN BETA CHAIN, FIBRINOGEN GAMMA CHAIN, ... | | Authors: | Macheboeuf, P, Y Fu, C, Zinkernagel, A.S, Johnson, J.E, Nizet, V, Ghosh, P. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Streptococcal M1 Protein Constructs a Pathological Host Fibrinogen Network
Nature, 472, 2011
|
|
2XVN
 
 | | A. fumigatus chitinase A1 phenyl-methylguanylurea complex | | Descriptor: | 1-METHYL-3-(N-PHENYLCARBAMIMIDOYL)UREA, ASPERGILLUS FUMIGATUS CHITINASE A1, CHLORIDE ION | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
1KXN
 
 | | Crystal Structure of Cytochrome c Peroxidase with a Proposed Electron Transfer Pathway Excised to Form a Ligand Binding Channel. | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome c peroxidase | | Authors: | Rosenfeld, R.J, Hayes, A.M.A, Musah, R.A, Goodin, D.B. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Excision of a proposed electron transfer pathway in cytochrome c peroxidase and its replacement by a ligand-binding channel.
Protein Sci., 11, 2002
|
|
1XXT
 
 | | The T-to-T High Transitions in Human Hemoglobin: wild-type deoxy Hb A (low salt, one test set) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-11-08 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
7DFY
 
 | | Novel motif for left-handed G-quadruplex formation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2xMotif2, POTASSIUM ION, ... | | Authors: | Das, P, Winnerdy, F.R, Maity, A, Mechulam, Y, Phan, A.T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A novel minimal motif for left-handed G-quadruplex formation.
Chem.Commun.(Camb.), 57, 2021
|
|
191L
 
 | |
2JYN
 
 | | A novel solution NMR structure of protein yst0336 from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium target YT51/Ontario Centre for Structural Proteomics target yst0336 | | Descriptor: | UPF0368 protein YPL225W | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Gutmanas, A, Semest, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel solution NMR structure of protein yst0336 from Saccharomyces cerevisiae.
To be Published
|
|
4KC9
 
 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
1L35
 
 | |
1C0Y
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
