1LXH
 
 | |
1M0L
 
 | | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.47 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal.
J.Mol.Biol., 321, 2002
|
|
1IDH
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
3CBF
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27 | | Descriptor: | (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]hexanedioic acid, Alpha-aminodipate aminotransferase | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-02-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
3D46
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate | | Descriptor: | L(+)-TARTARIC ACID, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate.
To be Published
|
|
3DZB
 
 | |
1LXG
 
 | |
3B74
 
 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylethanolamine | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Uncharacterized protein YKL091C | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
4PV6
 
 | | Crystal Structure Analysis of Ard1 from Thermoplasma volcanium | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-terminal acetyltransferase complex subunit [ARD1] | | Authors: | Ma, C, Lee, S.J, Lee, B.J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A.
Biochim.Biophys.Acta, 1844, 2014
|
|
4MLD
 
 | | X-ray structure of ComE D58E REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
3ELI
 
 | | Crystal structure of the AHSA1 (SPO3351) protein from Silicibacter pomeroyi, Northeast Structural Genomics Consortium Target SiR160 | | Descriptor: | Aha1 domain protein | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, D, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-22 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the AHSA1 (SPO3351) protein from Silicibacter pomeroyi, Northeast Structural Genomics Consortium Target SiR160
To be Published
|
|
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
3B20
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with NADfrom Synechococcus elongatus" | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Matsumura, H, Kai, A, Maeda, T, Inoue, T. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
1MF8
 
 | |
1NSL
 
 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
3D37
 
 | | The crystal structure of the tail protein from Neisseria meningitidis MC58 | | Descriptor: | CHLORIDE ION, Tail protein, 43 kDa | | Authors: | Zhang, R, Li, H, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the tail protein from Neisseria meningitidis MC58.
To be Published
|
|
1IDI
 
 | | THE NMR SOLUTION STRUCTURE OF ALPHA-BUNGAROTOXIN | | Descriptor: | ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
4PQ8
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
4PSH
 
 | | Structure of holo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3, ARGININE | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
1P5E
 
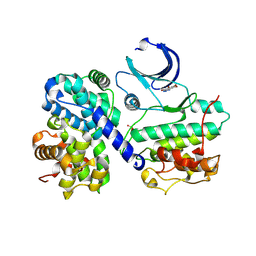 | | The structure of phospho-CDK2/cyclin A in complex with the inhibitor 4,5,6,7-tetrabromobenzotriazole (TBS) | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, Cell division protein kinase 2, Cyclin A2 | | Authors: | De Moliner, E, Brown, N.R, Johnson, L.N. | | Deposit date: | 2003-04-26 | | Release date: | 2003-07-01 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Alternative binding modes of an inhibitor to two different kinases
Eur.J.Biochem., 270, 2003
|
|
3EOA
 
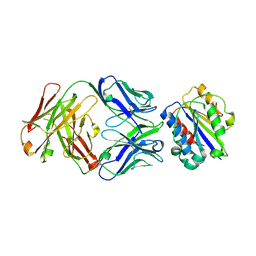 | |
1JV7
 
 | | BACTERIORHODOPSIN O-LIKE INTERMEDIATE STATE OF THE D85S MUTANT AT 2.25 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
1JGJ
 
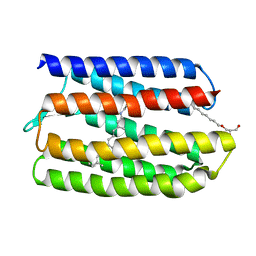 | |
4QO1
 
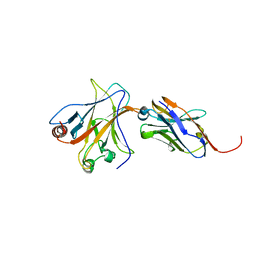 | | p53 DNA binding domain in complex with Nb139 | | Descriptor: | Cellular tumor antigen p53, Nb139 Nanobody against the DNA-binding domain of p53, ZINC ION | | Authors: | De Gieter, S, Bethuyne, J, Gettemans, J, Garcia-Pino, A, Loris, R. | | Deposit date: | 2014-06-19 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | A nanobody modulates the p53 transcriptional program without perturbing its functional architecture.
Nucleic Acids Res., 42, 2014
|
|
1JKG
 
 | | Structural basis for the recognition of a nucleoporin FG-repeat by the NTF2-like domain of TAP-p15 mRNA nuclear export factor | | Descriptor: | TAP, p15 | | Authors: | Fribourg, S, Braun, I.C, Izaurralde, E, Conti, E. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Mol.Cell, 8, 2001
|
|
