7C7Y
 
 | | C-terminal domain of B. cereus TubY | | Descriptor: | Uncharacterized protein | | Authors: | Hayashi, I. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-28 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C-terminal region of the plasmid partitioning protein TubY is a tetramer that can bind membranes and DNA.
J.Biol.Chem., 295, 2020
|
|
6AF2
 
 | | Crystal structure of N-terminus deletion mutant of Mycobacterium avium diadenosine 5',5'''-P1,P4-tetraphosphate phosphorylase | | Descriptor: | DI(HYDROXYETHYL)ETHER, HIT domain-containing protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Mori, S, Honda, N, Kim, H, Rimbara, E, Shibayama, K. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of N-terminus deletion mutant of Mycobacterium avium diadenosine tetraphosphate phosphorylase
To Be Published
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
8HK0
 
 | | Crystal structure of Fic32-33 complex from Streptomyces ficellus NRRL 8067 | | Descriptor: | Acyl-CoA dehydrogenase, Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cheng, Y, Qiao, H, Liu, W, Fang, P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
3QW4
 
 | | Structure of Leishmania donovani UMP synthase | | Descriptor: | CHLORIDE ION, UMP synthase, URIDINE-5'-MONOPHOSPHATE | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
4BO9
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 5-(2-(furan-2-ylmethoxy) phenyl)-2-phenyltetrazole at 2.9A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 5-[2-(FURAN-2-YLMETHOXY)PHENYL]-2-PHENYLTETRAZOLE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
1C3V
 
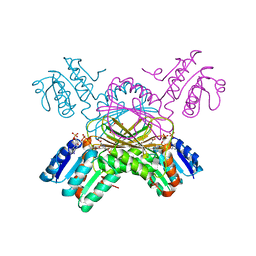 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
6YL7
 
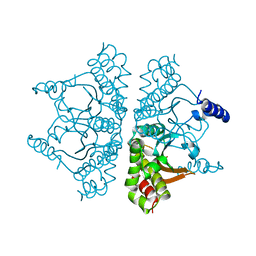 | |
8USW
 
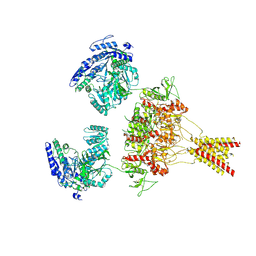 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
2WF3
 
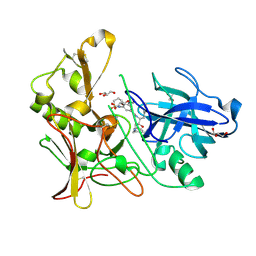 | | Human BACE-1 in complex with 6-(ethylamino)-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-1, 3,4,5-tetrahydro-2,1-benzothiazepine-8-carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, GLYCEROL, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-6-(ETHYLAMINO)-1-METHYL-1,3,4,5-TETRAHYDRO-2,1-BENZOTHIAZEPINE-8-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5TBP
 
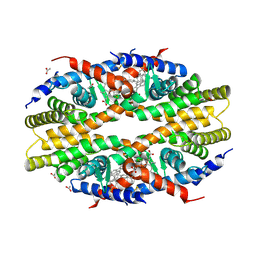 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
6YJN
 
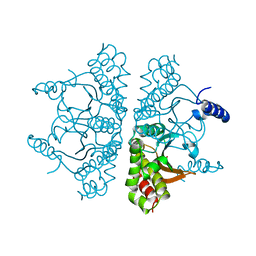 | |
5UHR
 
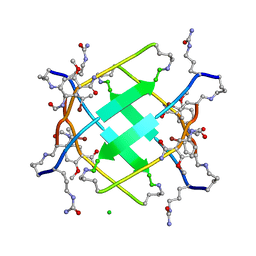 | |
7O62
 
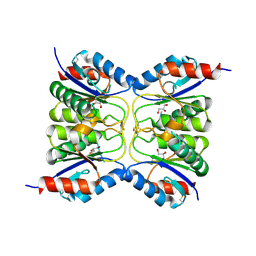 | |
5N5Q
 
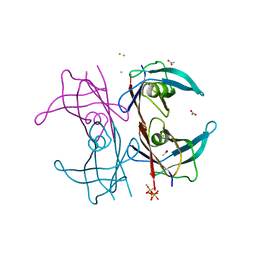 | | Human TTR altered conformation from soaking in iron chloride. | | Descriptor: | ACETATE ION, FE (III) ION, Transthyretin, ... | | Authors: | Ciccone, L, Savko, M, Shepard, W, Stura, E.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Copper mediated amyloid-beta binding to Transthyretin.
Sci Rep, 8, 2018
|
|
4AAY
 
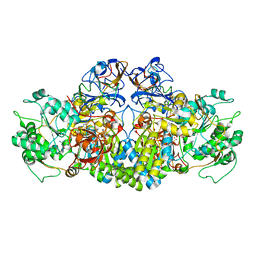 | | Crystal Structure of the arsenite oxidase protein complex from Rhizobium species strain NT-26 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, AROA, AROB, ... | | Authors: | Oke, M, Santini, J.M, Naismith, J.H. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Respiratory Arsenite Oxidase: Structure and the Role of Residues Surrounding the Rieske Cluster.
Plos One, 8, 2013
|
|
8JBB
 
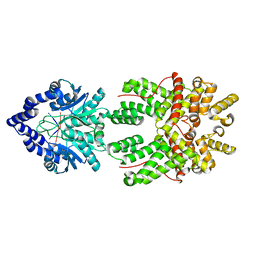 | |
2WF2
 
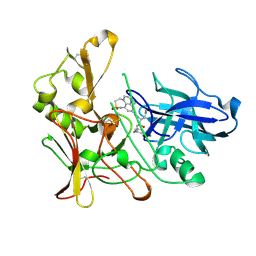 | | Human BACE-1 in complex with 8-ethyl-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-3,4,7, 8-tetrahydro-1H,6H-(1,2,5)thiadiazepino(5,4,3-de)quinoxaline-10- carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-8-ETHYL-1-METHYL-3,4,7,8-TETRAHYDRO-1H,6H-[1,2,5]THIADIAZEPINO[5,4,3-DE]QUINOXALINE-10-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5MZX
 
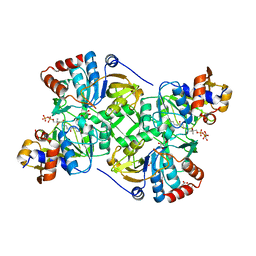 | | Crystal structure of the decarboxylase AibA/AibB in complex with 4'-diphospho pantetheine | | Descriptor: | 4'-diphospho pantetheine, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
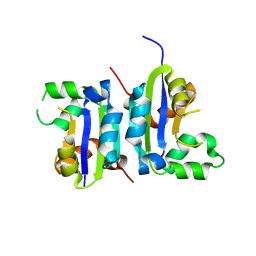
5X00
 
 | |
6WHV
 
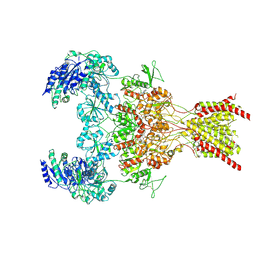 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WI0
 
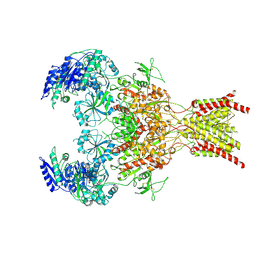 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
1XPH
 
 | |
6D3B
 
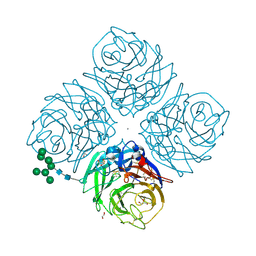 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) APO FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-04-15 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
