4D1T
 
 | | High resolution structure of native tVIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4D1V
 
 | | A F218Y mutant of VIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
2WOT
 
 | | ALK5 IN COMPLEX WITH 4-((5,6-dimethyl-2-(2-pyridyl)-3-pyridyl)oxy)-N-(3,4,5-trimethoxyphenyl)pyridin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5,6-DIMETHYL-2,2'-BIPYRIDIN-3-YL)OXY]-N-(3,4,5-TRIMETHOXYPHENYL)PYRIDIN-2-AMINE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Norman, R.A, Debreczeni, J.E, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
8CYI
 
 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | Authors: | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | Deposit date: | 2022-05-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
1L0X
 
 | |
1AE9
 
 | |
1L0Y
 
 | | T cell receptor beta chain complexed with superantigen SpeA soaked with zinc | | Descriptor: | 14.3.d T cell receptor beta chain, Exotoxin type A, GLYCEROL, ... | | Authors: | Li, H, Sundberg, E.J, Mariuzza, R.A. | | Deposit date: | 2002-02-14 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
Structure, 10, 2002
|
|
2WOU
 
 | | ALK5 IN COMPLEX WITH 4-((4-((2,6-dimethyl-3-pyridyl)oxy)-2-pyridyl) amino)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2,6-DIMETHYLPYRIDIN-3-YL)OXY]PYRIDIN-2-YL}AMINO)BENZENESULFONAMIDE, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Debreczeni, J.E, Norman, R.A, Goldberg, F.W, Ward, R.A, Finlay, R, Powell, S.J, Roberts, N.J, Dishington, A.P, Gingell, H.J, Wickson, K.F, Roberts, A.L. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Generation of a High Quality Lead for Transforming Growth Factor-Beta (Tgf-Beta) Type I Receptor (Alk5).
J.Med.Chem., 52, 2009
|
|
3KGF
 
 | | The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with phenylalanine and tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2009-10-29 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
7UM7
 
 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine | | Descriptor: | (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ1
 
 | | Crystal structure of PSF-RNA complex | | Descriptor: | RNA (30-MER), Splicing factor, proline- and glutamine-rich | | Authors: | Sachpatzidis, A, Wang, J, Konigsberg, W.H. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into the Tumor Suppression Mechanism from the Structure of Human Polypyrimidine Splicing Factor (PSF/SFPQ) Complexed with a 30mer RNA from Murine Virus-like 30S Transcript-1.
Biochemistry, 61, 2022
|
|
3G7W
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
3GC6
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-02-21 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3G7V
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
3GHH
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3E2L
 
 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
4CMP
 
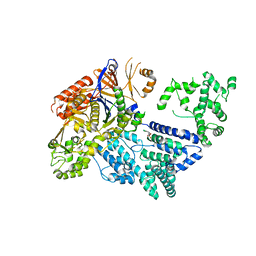 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
3GH3
 
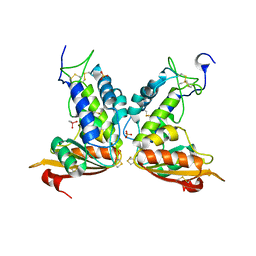 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, Ecto-NAD+ glycohydrolase (CD38 molecule), ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Kellenberger, E, Oppenheimer, N, Schuber, F. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
7Y9N
 
 | | an engineered 5-helix bundle derived from SARS-CoV-2 S2 in complex with HR2P | | Descriptor: | SARS-coV-2 S2 subunit, Spike protein S2',5HB-H2 | | Authors: | Lu, G.W, Lin, X, Guo, L.Y, Lin, S. | | Deposit date: | 2022-06-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | An engineered 5-helix bundle derived from SARS-CoV-2 S2 pre-binds sarbecoviral spike at both serological- and endosomal-pH to inhibit virus entry.
Emerg Microbes Infect, 11, 2022
|
|
7YKJ
 
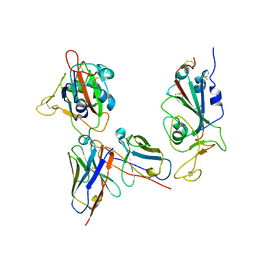 | | Omicron RBDs bound with P3E6 Fab (one up and one down) | | Descriptor: | P3E6 heavy chain, P3E6 light chain, Spike glycoprotein | | Authors: | Tang, B, Dang, S. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2.
Emerg Microbes Infect, 12, 2023
|
|
3HJT
 
 | |
3I0O
 
 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, in complex with ADP and Spectinomcyin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B, Berghuis, A.M. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
2JKE
 
 | | Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, ALPHA-GLUCOSIDASE (ALPHA-GLUCOSIDASE SUSB), ... | | Authors: | Gloster, T.M, Turkenburg, J.P, Potts, J.R, Henrissat, B, Davies, G.J. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergence of Catalytic Mechanism within a Glycosidase Family Provides Insight Into Evolution of Carbohydrate Metabolism by Human Gut Flora.
Chem.Biol., 15, 2008
|
|
1Y1W
 
 | | Complete RNA Polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*AP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
1HET
 
 | | atomic X-ray structure of liver alcohol dehydrogenase containing a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-25 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
