6CRM
 
 | |
6CZ5
 
 | | Crystal structure of small molecule AMP-acrylamide covalently bound to DDX3 S228C | | Descriptor: | 5'-O-[(R)-hydroxy(propanoylamino)phosphoryl]adenosine, ATP-dependent RNA helicase DDX3X | | Authors: | Barkovich, K.J, Moore, M.K, Hu, Q, Shokat, K.M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemical genetic inhibition of DEAD-box proteins using covalent complementarity.
Nucleic Acids Res., 46, 2018
|
|
6D6R
 
 | |
6D6Q
 
 | |
6DCR
 
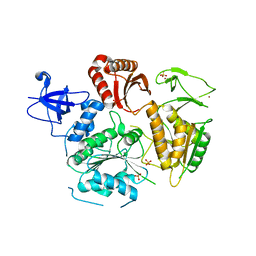 | |
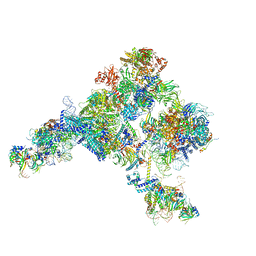
5ZWO
 
 | | Cryo-EM structure of the yeast B complex at average resolution of 3.9 angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | Authors: | Bai, R, Wan, R, Yan, C, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
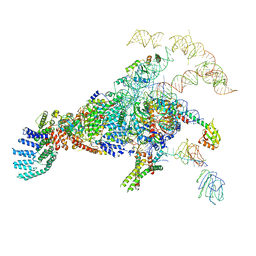
5ZWM
 
 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.4~4.6 angstrom (tri-snRNP and U2 snRNP Part) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5ZWN
 
 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.3 angstrom (Part II: U1 snRNP region) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Pre-mRNA-splicing ATP-dependent RNA helicase PRP28, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
6DGD
 
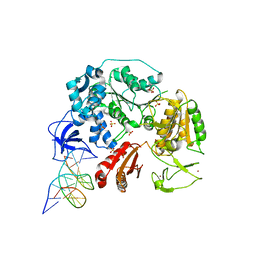 | | PriA helicase bound to dsDNA of a DNA replication fork | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*CP*CP*GP*AP*CP*T)-3'), DNA (5'-D(P*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*AP*CP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Satyshur, K.A, Windgassen, T.A, Keck, J.L. | | Deposit date: | 2018-05-17 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.823 Å) | | Cite: | Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GPG
 
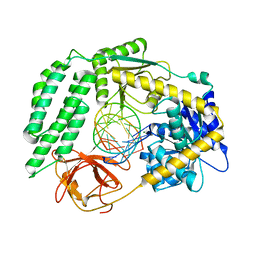 | | Structure of the RIG-I Singleton-Merten syndrome variant C268F | | Descriptor: | MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*CP*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*CP*G)-3'), ... | | Authors: | Laessig, C, Lammens, K, Hopfner, K.-P. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Unified mechanisms for self-RNA recognition by RIG-I Singleton-Merten syndrome variants.
Elife, 7, 2018
|
|
6H57
 
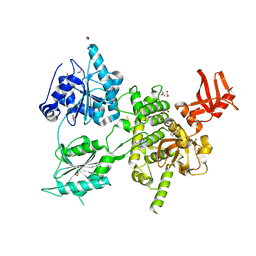 | | Crystal structure of S. cerevisiae DEAH-box RNA helicase Dhr1, essential for small ribosomal subunit biogenesis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Roychowdhury, A, Graille, M. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The DEAH-box RNA helicase Dhr1 contains a remarkable carboxyl terminal domain essential for small ribosomal subunit biogenesis.
Nucleic Acids Res., 47, 2019
|
|
6AC6
 
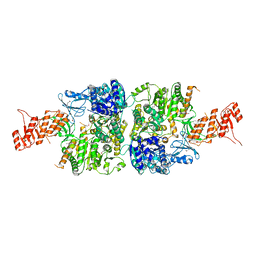 | | Ab initio crystal structure of Selenomethionine labelled Mycobacterium smegmatis Mfd | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6AC8
 
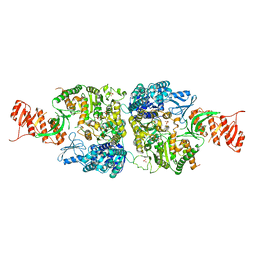 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACA
 
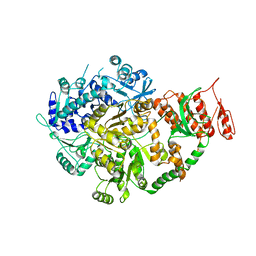 | | Crystal structure of Mycobacterium tuberculosis Mfd at 3.6 A resolution | | Descriptor: | Mycobacterium tuberculosis Mfd | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACX
 
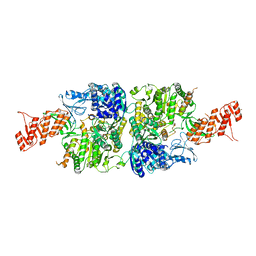 | | Crystal structure of Mycobacterium smegmatis Mfd in complex with ADP + Pi at 3.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mycobacterium smegmatis Mfd, PHOSPHATE ION, ... | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6AH0
 
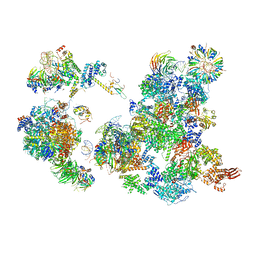 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6AHD
 
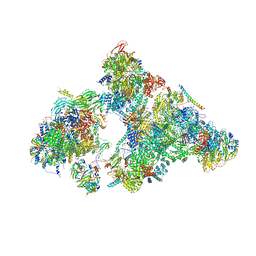 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6HEG
 
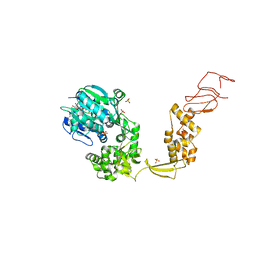 | | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpB, PHOSPHATE ION, ... | | Authors: | Xin, B.G, Chen, W.F, Rety, S, Dai, Y.X, Xi, X.G. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AIB
 
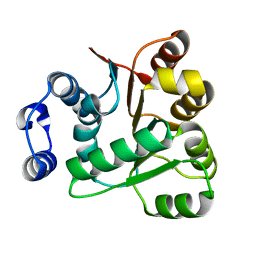 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6AIC
 
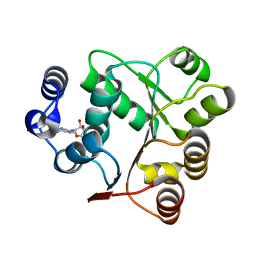 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6IEG
 
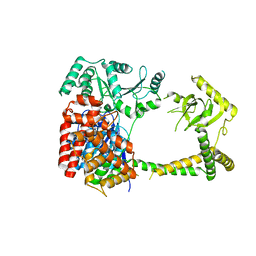 | | Crystal structure of human MTR4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6IEH
 
 | | Crystal structures of the hMTR4-NRDE2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6HYT
 
 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.3 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
