8DMB
 
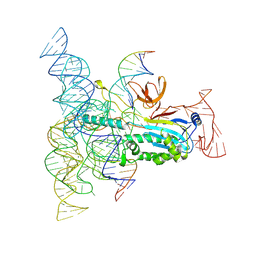 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
8E7O
 
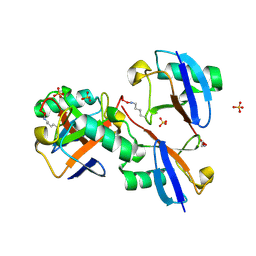 | | CRYSTAL STRUCTURE OF LYS48-LINKED TETRAUBIQUITIN | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Lemma, B.E, Fushman, D. | | Deposit date: | 2022-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
8DMS
 
 | |
8DMQ
 
 | |
5VSX
 
 | |
5W46
 
 | |
5X3O
 
 | |
5X3N
 
 | |
5XIT
 
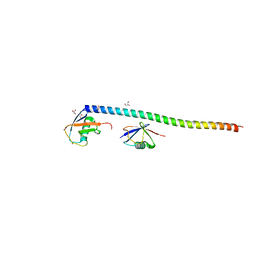 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form II | | Descriptor: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XDP
 
 | |
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XPK
 
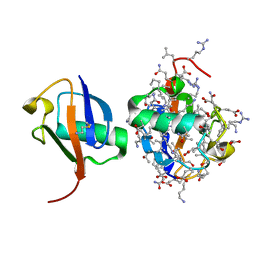 | |
5X3M
 
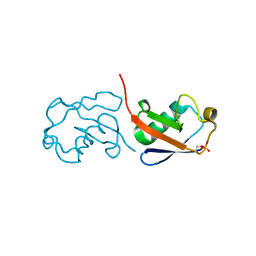 | |
5XIU
 
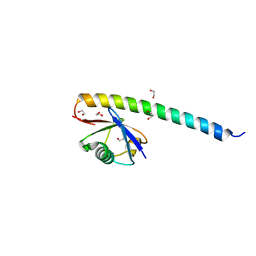 | | Crystal structure of RNF168 UDM2 in complex with Lys63-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF168, Ubiquitin-40S ribosomal protein S27a | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XK4
 
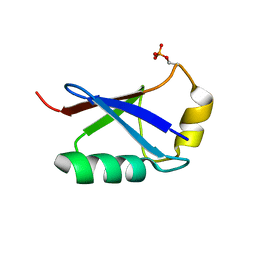 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XK5
 
 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YC2
 
 | |
5VF0
 
 | |
5YDK
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, tetrameric form | | Descriptor: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168
Nat Commun, 9, 2018
|
|
5YCA
 
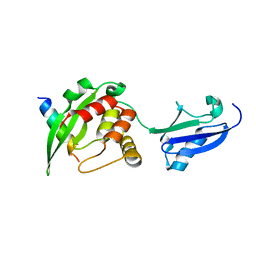 | | Crystal structure of inner membrane protein Bqt4 in complex with LEM2 | | Descriptor: | Lap-Emerin-Man domain protein 2, Ubiquitin-like protein SMT3,Bouquet formation protein 4 | | Authors: | Chen, Y, Hu, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into chromosome attachment to the nuclear envelope by an inner nuclear membrane protein Bqt4 in fission yeast.
Nucleic Acids Res., 47, 2019
|
|
5VSZ
 
 | |
5XQM
 
 | |
5ZEW
 
 | |
6GF2
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
6GF1
 
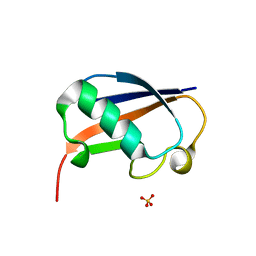 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | SULFATE ION, Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
