3OVE
 
 | |
3S8L
 
 | |
3S8O
 
 | |
3OV1
 
 | |
3T04
 
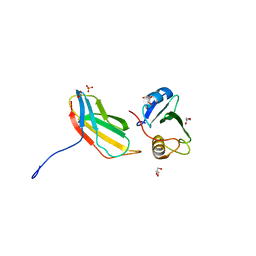 | | Crystal structure of monobody 7c12/abl1 sh2 domain complex | | Descriptor: | GLYCEROL, MONOBODY 7C12, SULFATE ION, ... | | Authors: | Wojcik, J.B, Wyrzucki, A.M, Koide, S. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the SH2-Kinase Interface in Bcr-Abl Inhibits Leukemogenesis.
Cell(Cambridge,Mass.), 147, 2011
|
|
3UF4
 
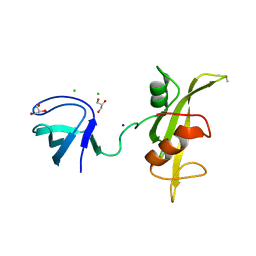 | |
4A55
 
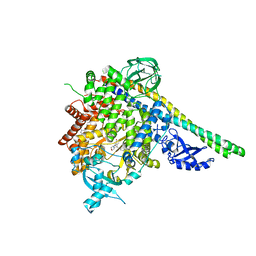 | | Crystal structure of p110alpha in complex with iSH2 of p85alpha and the inhibitor PIK-108 | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM, PIK-108 | | Authors: | Hon, W.-C, Berndt, A, Williams, R.L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Regulation of Lipid Binding Underlies the Activation Mechanism of Class Ia Pi3-Kinases.
Oncogene, 31, 2012
|
|
3US4
 
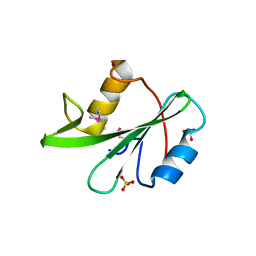 | |
3UYO
 
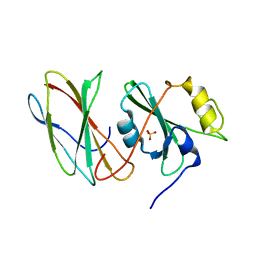 | |
4D8K
 
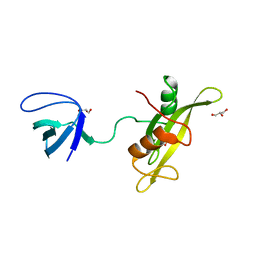 | |
3S9K
 
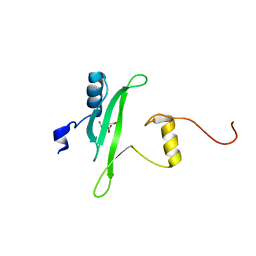 | | Crystal structure of the Itk SH2 domain. | | Descriptor: | CITRIC ACID, Tyrosine-protein kinase ITK/TSK | | Authors: | Joseph, R.E, Ginder, N.D, Hoy, J.A, Nix, J.C, Fulton, B.D, Honzatko, R.B, Andreotti, A.H. | | Deposit date: | 2011-06-01 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structure of the interleukin-2 tyrosine kinase Src homology 2 domain; comparison between X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E93
 
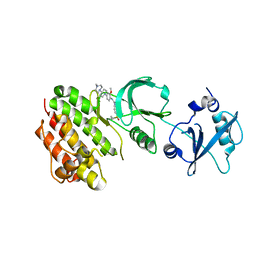 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES)in complex with TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, Tyrosine-protein kinase Fes/Fps | | Authors: | Filippakopoulos, P, Salah, E, Miduturu, C.V, Fedorov, O, Cooper, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Small-Molecule Inhibitors of the c-Fes Protein-Tyrosine Kinase.
Chem.Biol., 19, 2012
|
|
2LQN
 
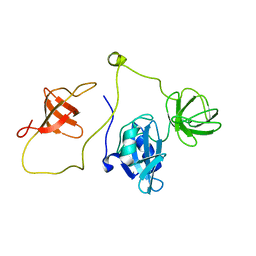 | | Solution structure of CRKL | | Descriptor: | Crk-like protein | | Authors: | Jankowski, W, Saleh, T, Kalodimos, C. | | Deposit date: | 2012-03-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain organization differences explain Bcr-Abl's preference for CrkL over CrkII.
Nat.Chem.Biol., 8, 2012
|
|
2LQW
 
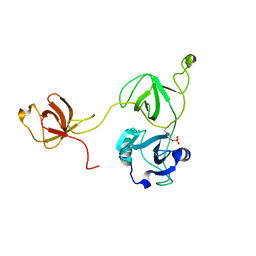 | |
4F5A
 
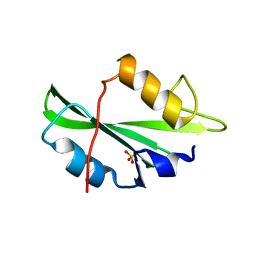 | | Triple mutant Src SH2 domain bound to phosphate ion | | Descriptor: | PHOSPHATE ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F59
 
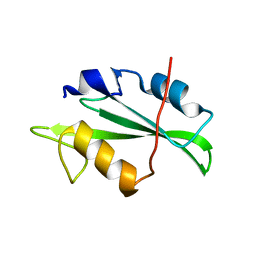 | | Triple mutant Src SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F5B
 
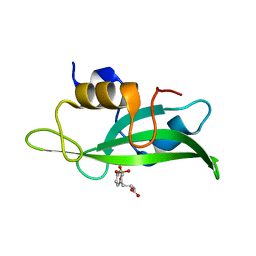 | | Triple mutant Src SH2 domain bound to phosphotyrosine | | Descriptor: | O-PHOSPHOTYROSINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4EY0
 
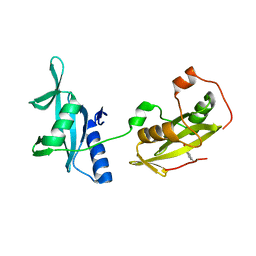 | | Structure of tandem SH2 domains from PLCgamma1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
4FBN
 
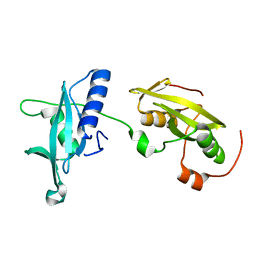 | | Insights into structural integration of the PLCgamma regulatory region and mechanism of autoinhibition and activation based on key roles of SH2 domains | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
2LNW
 
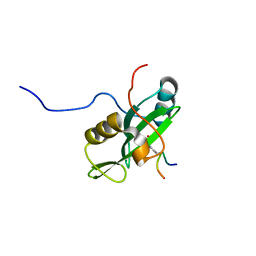 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
 | | Solution structure of Vav2 SH2 domain | | Descriptor: | Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
4FL2
 
 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
4FL3
 
 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
4E68
 
 | | Unphosphorylated STAT3B core protein binding to dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Collie, G.W, Parkinson, G.N, Shah, R. | | Deposit date: | 2012-03-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography.
Febs Lett., 587, 2013
|
|
4DGP
 
 | | The wild-type Src homology 2 (SH2)-domain containing protein tyrosine phosphatase-2 (SHP2) | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-26 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
