2I83
 
 | | hyaluronan-binding domain of CD44 in its ligand-bound form | | Descriptor: | CD44 antigen | | Authors: | Takeda, M, Ogino, S, Umemoto, R, Sakakura, M, Kajiwara, M, Sugahara, K.N, Hayasaka, H, Miyasaka, M, Terasawa, H, Shimada, I. | | Deposit date: | 2006-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Ligand-induced Structural Changes of the CD44 Hyaluronan-binding Domain Revealed by NMR
J.Biol.Chem., 281, 2006
|
|
2JUJ
 
 | |
2K0J
 
 | | Solution structure of CaM complexed to DRP1p | | Descriptor: | CALCIUM ION, LANTHANUM (III) ION, calmodulin | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-02-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
2M0D
 
 | |
2JNY
 
 | | Solution NMR structure of protein Uncharacterized BCR, Northeast Structural Genomics Consortium target CgR1 | | Descriptor: | Uncharacterized BCR | | Authors: | Wu, Y, Liu, G, Zhang, Q, Chen, C, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M, Swapna, G, Acton, T, Rost, B, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Uncharacterized BCR, Northeast Structural Genomics Consortium target CgR1
To be Published
|
|
2LVZ
 
 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
2MAZ
 
 | |
2LCX
 
 | |
2N0J
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | Descriptor: | RIBOSTAMYCIN, RNA_(27-MER) | | Authors: | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | Deposit date: | 2015-03-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2ABY
 
 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
2PCF
 
 | | THE COMPLEX OF CYTOCHROME F AND PLASTOCYANIN DETERMINED WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | COPPER (II) ION, CYTOCHROME F, HEME C, ... | | Authors: | Ubbink, M, Ejdeback, M, Karlsson, B.G, Bendall, D.S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics.
Structure, 6, 1998
|
|
2LT3
 
 | | Solution NMR structure of the C-terminal domain of CdnL from Myxococcus xanthus | | Descriptor: | Transcriptional regulator, CarD family | | Authors: | Mirassou, Y, Garcia-Moreno, D, Padmanabhan, S, Jimenez, M.A. | | Deposit date: | 2012-05-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into RNA Polymerase Recognition and Essential Function of Myxococcus xanthus CdnL.
Plos One, 9, 2014
|
|
1WTU
 
 | | TRANSCRIPTION FACTOR 1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSCRIPTION FACTOR 1 | | Authors: | Jia, X, Grove, A, Ivancic, M, Hsu, V.L, Geiduschek, E.P, Kearns, D.R. | | Deposit date: | 1996-07-29 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bacillus subtilis phage SPO1-encoded type II DNA-binding protein TF1 in solution.
J.Mol.Biol., 263, 1996
|
|
2M7M
 
 | | N-terminal domain of EhCaBP1 structure | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Chary, K.V, Rout, A.K, Patel, S, Bhattacharya, A. | | Deposit date: | 2013-04-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional Manipulation of a Calcium-binding Protein from Entamoeba histolytica Guided by Paramagnetic NMR.
J.Biol.Chem., 288, 2013
|
|
1VND
 
 | | VND/NK-2 PROTEIN (HOMEODOMAIN), NMR | | Descriptor: | VND/NK-2 PROTEIN | | Authors: | Tsao, D.H.H, Gruschus, J.M, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1996-05-22 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the NK-2 homeodomain from Drosophila.
J.Mol.Biol., 251, 1995
|
|
2M7N
 
 | | C-terminal structure of (Y81F)-EhCaBP1 | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Chary, K.V, Rout, A.K, Patel, S, Bhattacharya, A. | | Deposit date: | 2013-04-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional Manipulation of a Calcium-binding Protein from Entamoeba histolytica Guided by Paramagnetic NMR.
J.Biol.Chem., 288, 2013
|
|
2MUI
 
 | |
2CPS
 
 | | SOLUTION NMR STRUCTURES OF THE MAJOR COAT PROTEIN OF FILAMENTOUS BACTERIOPHAGE M13 SOLUBILIZED IN SODIUM DODECYL SULPHATE MICELLES, 25 LOWEST ENERGY STRUCTURES | | Descriptor: | M13 MAJOR COAT PROTEIN | | Authors: | Papavoine, C.H.M, Christiaans, B.E.C, Folmer, R.H.A, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
J.Mol.Biol., 282, 1998
|
|
2CPB
 
 | | SOLUTION NMR STRUCTURES OF THE MAJOR COAT PROTEIN OF FILAMENTOUS BACTERIOPHAGE M13 SOLUBILIZED IN DODECYLPHOSPHOCHOLINE MICELLES, 25 LOWEST ENERGY STRUCTURES | | Descriptor: | M13 MAJOR COAT PROTEIN | | Authors: | Papavoine, C.H.M, Christiaans, B.E.C, Folmer, R.H.A, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
J.Mol.Biol., 282, 1998
|
|
1XYX
 
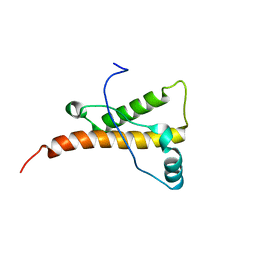 | | mouse prion protein fragment 121-231 | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2N8R
 
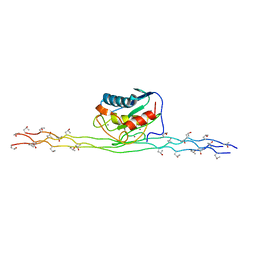 | |
2MOB
 
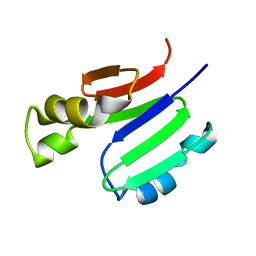 | | METHANE MONOOXYGENASE COMPONENT B | | Descriptor: | PROTEIN (METHANE MONOOXYGENASE REGULATORY PROTEIN B) | | Authors: | Chang, S.L, Wallar, B.J, Lipscomb, J.D, Mayo, K.H. | | Deposit date: | 1999-03-10 | | Release date: | 1999-08-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of component B from methane monooxygenase derived through heteronuclear NMR and molecular modeling.
Biochemistry, 38, 1999
|
|
2MIQ
 
 | | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 of Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J | | Descriptor: | Lysine-specific demethylase lid, ZINC ION | | Authors: | Xu, X, Eletsky, A, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 from Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J
To be Published
|
|
2MOA
 
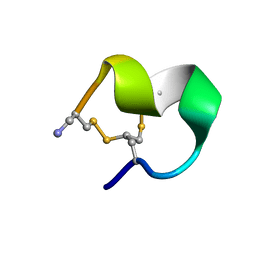 | |
1XSW
 
 | | The solid-state NMR structure of Kaliotoxin | | Descriptor: | Kaliotoxin 1 | | Authors: | Lange, A, Becker, S, Seidel, K, Giller, K, Pongs, O, Baldus, M. | | Deposit date: | 2004-10-20 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | A Concept for Rapid Protein-Structure Determination by Solid-State NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
