1C0N
 
 | | CSDB PROTEIN, NIFS HOMOLOGUE | | Descriptor: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | Deposit date: | 1999-07-17 | | Release date: | 2000-07-17 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
2O0T
 
 | |
3S9B
 
 | |
2O7E
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides (His89Phe variant), bound to 2-aminoindan-2-phosphonic acid | | Descriptor: | (2-AMINO-2,3-DIHYDRO-1H-INDEN-2-YL)PHOSPHONIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-11 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
1GZI
 
 | | CRYSTAL STRUCTURE OF TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT, AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN | | Authors: | Antson, A.A, Lewis, S, Roper, D.I, Smith, D.J, Hubbard, R.E. | | Deposit date: | 1996-10-25 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Type III Antifreeze Protein from Ocean Pout
To be Published
|
|
1YDU
 
 | | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain | | Descriptor: | At5g01610 | | Authors: | Zhao, Q, Cornilescu, C.C, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-26 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain
To be Published
|
|
3U69
 
 | | Unliganded wild-type human thrombin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Figueiredo, A.C, Clement, C.C, Philipp, M, Barbosa Pereira, P.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational design and characterization of d-phe-pro-d-arg-derived direct thrombin inhibitors.
Plos One, 7, 2012
|
|
2C1M
 
 | | Nup50:importin-alpha complex | | Descriptor: | IMPORTIN-ALPHA2 SUBUNIT, NUCLEOPORIN 50 KDA | | Authors: | Matsuura, Y, Stewart, M. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nup50/Npap60 Function in Nuclear Protein Import Complex Disassembly and Importin Recycling.
Embo J., 24, 2005
|
|
1GZA
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
1YC2
 
 | | Sir2Af2-NAD-ADPribose-nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase 2, ... | | Authors: | Avalos, J.L, Bever, M.K, Wolberger, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD(+) Cosubstrate Specificity of a Sir2 Enzyme.
Mol.Cell, 17, 2005
|
|
1CF9
 
 | |
1CI8
 
 | | ESTERASE ESTB FROM BURKHOLDERIA GLADIOLI: AN ESTERASE WITH (BETA)-LACTAMASE FOLD. | | Descriptor: | ISOPROPYL ALCOHOL, PROTEIN (CARBOXYLESTERASE) | | Authors: | Wagner, U.G, Petersen, E.I, Schwab, H, Kratky, C. | | Deposit date: | 1999-04-08 | | Release date: | 2001-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | EstB from Burkholderia gladioli: a novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity
Protein Sci., 11, 2002
|
|
1YEL
 
 | | Structure of the hypothetical Arabidopsis thaliana protein At1g16640.1 | | Descriptor: | At1g16640 | | Authors: | Peterson, F.C, Waltner, J.K, Lytle, B.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the B3 domain from Arabidopsis thaliana protein At1g16640
Protein Sci., 14, 2005
|
|
1FOH
 
 | | PHENOL HYDROXYLASE FROM TRICHOSPORON CUTANEUM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, PHENOL HYDROXYLASE | | Authors: | Enroth, C, Neujahr, H, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-03-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
Structure, 6, 1998
|
|
1XD2
 
 | | Crystal Structure of a ternary Ras:SOS:Ras*GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
3TTA
 
 | |
1CEB
 
 | |
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | Descriptor: | BROMIDE ION, putative acetyltransferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
3QPZ
 
 | | Crystal structure of the N59A mutant of the 3-deoxy-d-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J, Cochrane, F.C. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Targeting the role of a key conserved motif for substrate selection and catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase
Biochemistry, 50, 2011
|
|
1XFG
 
 | | Glutaminase domain of glucosamine 6-phosphate synthase complexed with l-glu hydroxamate | | Descriptor: | ACETATE ION, GLUTAMINE HYDROXAMATE, Glucosamine--fructose-6-phosphate aminotransferase [isomerizing], ... | | Authors: | Isupov, M.N, Teplyakov, A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Binding is Required for Assembly of the Active Conformation of the Catalytic Site in Ntn Amidotransferases: Evidence from the 1.8 Angstrom Crystal Structure of the Glutaminase Domain of Glucosamine 6-Phosphate Synthase
Structure, 4, 1996
|
|
2Q1W
 
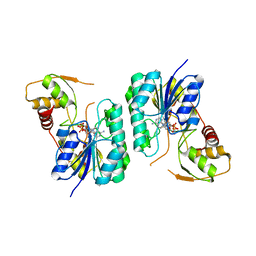 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmH in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | King, J.D, Harmer, N.J, Maskell, D.J, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
1XMB
 
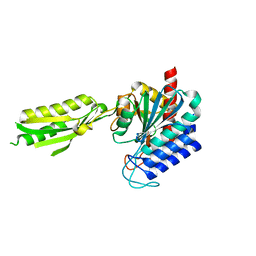 | | X-ray structure of IAA-aminoacid hydrolase from Arabidopsis thaliana gene AT5G56660 | | Descriptor: | IAA-amino acid hydrolase homolog 2 | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana.
Proteins, 74, 2009
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
1FAX
 
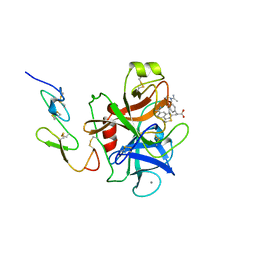 | | COAGULATION FACTOR XA INHIBITOR COMPLEX | | Descriptor: | (2S)-3-(7-carbamimidoylnaphthalen-2-yl)-2-[4-({(3R)-1-[(1Z)-ethanimidoyl]pyrrolidin-3-yl}oxy)phenyl]propanoic acid, CALCIUM ION, FACTOR XA | | Authors: | Brandstetter, H, Engh, R.A. | | Deposit date: | 1996-08-23 | | Release date: | 1997-10-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of active site-inhibited clotting factor Xa. Implications for drug design and substrate recognition.
J.Biol.Chem., 271, 1996
|
|
3QCM
 
 | |
