1DC3
 
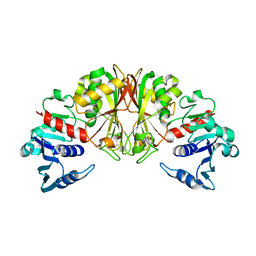 | |
4RWJ
 
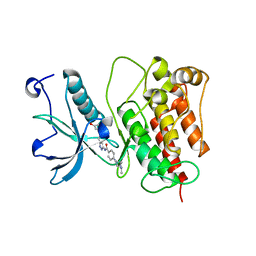 | | Crystal Structure of FGFR1 (C488A, C584S) in complex with AZD4547 (N-{3-[2-(3,5-DIMETHOXYPHENYL)ETHYL]-1H-PYRAZOL-5-YL}-4-[(3R,5S)-3,5-DIMETHYLPIPERAZIN-1-YL]BENZAMIDE) | | Descriptor: | Fibroblast growth factor receptor 1, N-{3-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]benzamide | | Authors: | Sohl, C.D, Anderson, K.S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Illuminating the Molecular Mechanisms of Tyrosine Kinase Inhibitor Resistance for the FGFR1 Gatekeeper Mutation: The Achilles' Heel of Targeted Therapy.
Acs Chem.Biol., 10, 2015
|
|
1DDX
 
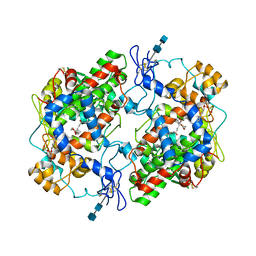 | | CRYSTAL STRUCTURE OF A MIXTURE OF ARACHIDONIC ACID AND PROSTAGLANDIN BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2: PROSTAGLANDIN STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[6-(3-HYDROPEROXY-OCT-1-ENYL)-2,3-DIOXA-BICYCLO[2.2.1]HEPT-5-YL]-HEPT-5-ENOIC ACID, ... | | Authors: | Kiefer, J.R, Pawlitz, J.L, Moreland, K.T, Stegeman, R.A, Gierse, J.K, Stevens, A.M, Goodwin, D.C, Rowlinson, S.W, Marnett, L.J, Stallings, W.C, Kurumbail, R.G. | | Deposit date: | 1999-11-11 | | Release date: | 2000-05-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the stereochemistry of the cyclooxygenase reaction.
Nature, 405, 2000
|
|
1IWJ
 
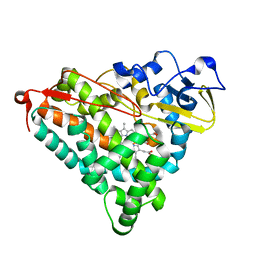 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(109K) Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
1IWW
 
 | | Crystal Structure Analysis of Human lysozyme at 152K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2BCN
 
 | |
1J0H
 
 | |
1J0X
 
 | | Crystal structure of the rabbit muscle glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Cowan-Jacob, S.W, Kaufmann, M, Anselmo, A.N, Stark, W, Grutter, M.G. | | Deposit date: | 2002-11-25 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1DHR
 
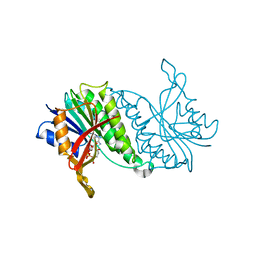 | | CRYSTAL STRUCTURE OF RAT LIVER DIHYDROPTERIDINE REDUCTASE | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Skinner, M.M, Whiteley, J.M, Matthews, D.A, Xuong, N.H. | | Deposit date: | 1992-03-30 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rat liver dihydropteridine reductase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
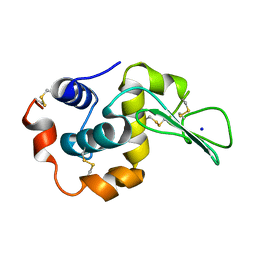
1DI5
 
 | |
3FQM
 
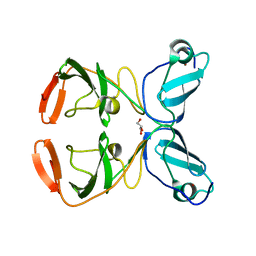 | |
6INR
 
 | | The crystal structure of phytoplasmal effector causing phyllody symptoms 1 (PHYL1) | | Descriptor: | CADMIUM ION, Putative effector, AYWB SAP54-like protein | | Authors: | Liao, Y.T, Lin, S.S, Ko, T.P, Wang, H.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the interaction between phytoplasmal effector causing phyllody 1 and MADS transcription factors.
Plant J., 100, 2019
|
|
6EHT
 
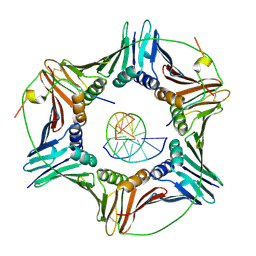 | | Modulation of PCNA sliding surface by p15PAF suggests a suppressive mechanism for cisplatin-induced DNA lesion bypass by pol eta holoenzyme | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), PCNA-associated factor, ... | | Authors: | De March, M, Barrera-Vilarmau, S, Mentegari, E, Merino, N, Bressan, E, Maga, G, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
1OYI
 
 | | Solution structure of the Z-DNA binding domain of the vaccinia virus gene E3L | | Descriptor: | double-stranded RNA-binding protein | | Authors: | Kahmann, J.D, Wecking, D.A, Putter, V, Lowenhaupt, K, Kim, Y.-G, Schmieder, P, Oschkinat, H, Rich, A, Schade, M. | | Deposit date: | 2003-04-04 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1J51
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F/V247L/C334A) WITH 1,3,5-TRICHLOROBENZENE | | Descriptor: | 1,3,5-TRICHLORO-BENZENE, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Chen, X, Christopher, A, Jones, J, Guo, Q, Xu, F, Cao, R, Wong, L.L, Rao, Z. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the F87W/Y96F/V247L mutant of cytochrome P-450cam with 1,3,5-trichlorobenzene bound and further protein engineering for the oxidation of pentachlorobenzene and hexachlorobenzene
J.BIOL.CHEM., 277, 2002
|
|
1P0H
 
 | | Crystal Structure of Rv0819 from Mycobacterium Tuberculosis MshD-Mycothiol Synthase Coenzyme A Complex | | Descriptor: | ACETYL COENZYME *A, COENZYME A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
4NFA
 
 | | Structure of the C-terminal doamin of Knl1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein CASC5 | | Authors: | Petrovic, A, Mosalaganti, S, Keller, J, Mattiuzzo, M, Overlack, K, Wohlgemuth, S, Pasqualato, S, Raunser, S, Musacchio, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization.
Mol.Cell, 53, 2014
|
|
1DLM
 
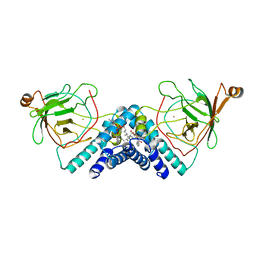 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER CALCOACETICUS NATIVE DATA | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, [1-PENTADECANOYL-2-DECANOYL-GLYCEROL-3-YL]PHOSPHONYL CHOLINE | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
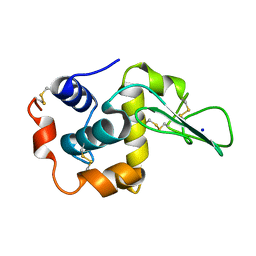
1DI3
 
 | |
4ZW2
 
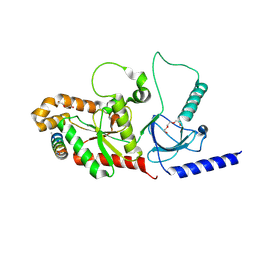 | |
1IEW
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1DC4
 
 | | STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Yun, M, Park, C.-G, Kim, J.-Y, Park, H.-W. | | Deposit date: | 1999-11-04 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes.
Biochemistry, 39, 2000
|
|
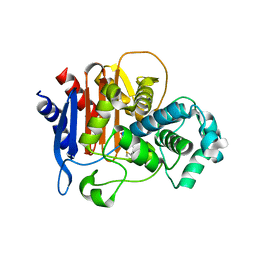
5W13
 
 | |
1II5
 
 | |
4R4M
 
 | | Crystal structure of C42L cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
