1WA2
 
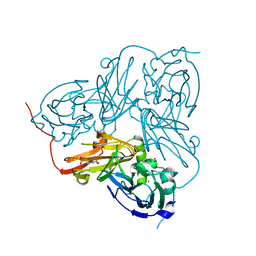 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase with nitrite bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE,, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA3
 
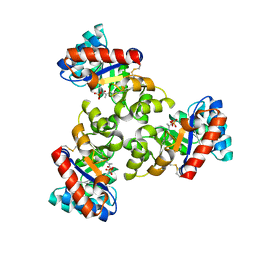 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1WA4
 
 | |
1WA5
 
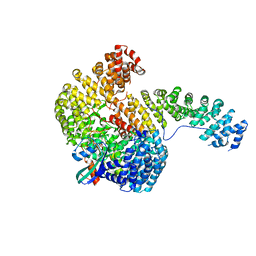 | | Structure of the Cse1:Imp-alpha:RanGTP complex | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin alpha re-exporter, ... | | Authors: | Stewart, M. | | Deposit date: | 2004-10-23 | | Release date: | 2004-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the assembly of a nuclear export complex.
Nature, 432, 2004
|
|
1WA6
 
 | | The structure of ACC oxidase | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 1, FE (II) ION, PHOSPHATE ION, ... | | Authors: | Zhang, Z, Ren, J.-S, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2004-10-25 | | Release date: | 2005-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure and Mechanistic Implications of 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (the Ethyling Forming Enzyme)
Chem.Biol., 11, 2004
|
|
1WA7
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE IN COMPLEX WITH A HERPESVIRAL LIGAND | | Descriptor: | HYPOTHETICAL 28.7 KDA PROTEIN IN DHFR 3'REGION (ORF1), TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-10-25 | | Release date: | 2005-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck.
Biochemistry, 41, 2002
|
|
1WA8
 
 | | Solution Structure of the CFP-10.ESAT-6 Complex. Major Virulence Determinants of Pathogenic Mycobacteria | | Descriptor: | 6 KDA EARLY SECRETORY ANTIGENIC TARGET (ESAT-6), ESAT-6 LIKE PROTEIN ESXB | | Authors: | Renshaw, P.S, Lightbody, K.L, Veverka, V, Muskett, F.W, Kelly, G, Frenkiel, T.A, Gordon, S.V, Hewinson, R.G, Burke, B, Norman, J, Williamson, R.A, Carr, M.D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-25 | | Release date: | 2005-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of the Complex Formed by the Tuberculosis Virulence Factors Cfp-10 and Esat-6
Embo J., 24, 2005
|
|
1WA9
 
 | | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | | Descriptor: | PERIOD CIRCADIAN PROTEIN | | Authors: | Yildiz, O, Doi, M, Yujnovsky, I, Cardone, L, Berndt, A, Hennig, S, Schulze, S, Urbanke, C, Sassone-Corsi, P, Wolf, E. | | Deposit date: | 2004-10-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Mol.Cell, 17, 2005
|
|
1WAA
 
 | | IG27 protein domain | | Descriptor: | TITIN, ZINC ION | | Authors: | Vega, M.C, Valencia, L, Zou, P, Wilmanns, M. | | Deposit date: | 2004-10-25 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanical Network in Titin Immunoglobulin from Force Distribution Analysis.
Plos Comput.Biol., 5, 2009
|
|
1WAB
 
 | | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | ACETATE ION, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Swenson, L, Derewenda, U, Serre, L, Wei, Y, Dauter, Z, Hattori, M, Aoki, J, Arai, H, Adachi, T, Inoue, K, Derewenda, Z.S. | | Deposit date: | 1996-10-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer.
Nature, 385, 1997
|
|
1WAC
 
 | | Back-priming mode of Phi6 RNA-dependent RNA polymerase | | Descriptor: | P2 PROTEIN | | Authors: | Laurila, M.R.L, Salgado, P.S, Stuart, D.I, Grimes, J.M, Bamford, D.H. | | Deposit date: | 2004-10-26 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Back-Priming Mode of Phi6 RNA-Dependent RNA Polymerase
J.Gen.Virol., 86, 2005
|
|
1WAD
 
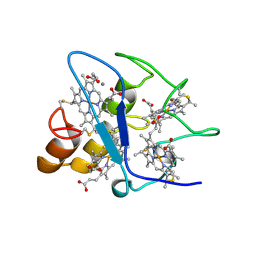 | | CYTOCHROME C3 WITH 4 HEME GROUPS AND ONE CALCIUM ION | | Descriptor: | CALCIUM ION, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Morais, J, Coelho, R, Carrondo, M.A, Wilson, K, Dauter, Z, Sieker, L. | | Deposit date: | 1996-01-10 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytochrome c3 from Desulfovibrio gigas: crystal structure at 1.8 A resolution and evidence for a specific calcium-binding site.
Protein Sci., 5, 1996
|
|
1WAE
 
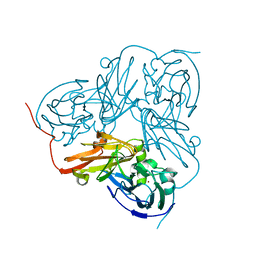 | | Crystal structure of H129V Mutant of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2004-10-26 | | Release date: | 2004-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observation of an Unprecedented Cu Bis-His Site: Crystal Structure of the H129V Mutant of Nitrite Reductase
Inorg.Chem., 43, 2004
|
|
1WAF
 
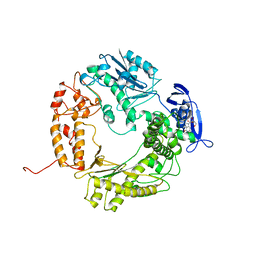 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
1WAJ
 
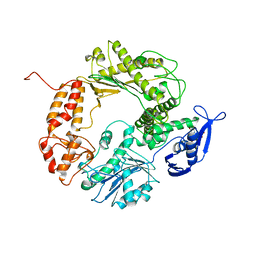 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
1WAK
 
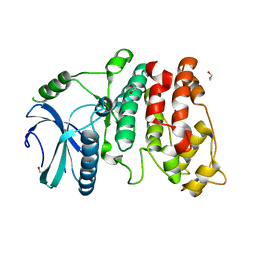 | | X-ray structure of SRPK1 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE SPRK1 | | Authors: | Ngo, J.C, Gullingsrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.D, Adams, J.A, Mccammon, J.A, Ghosh, G. | | Deposit date: | 2004-10-26 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Sr Protein Kinase 1 is Resilient to Inactivation.
Structure, 15, 2007
|
|
1WAL
 
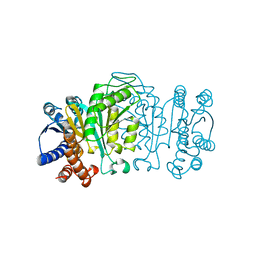 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) MUTANT (M219A)FROM THERMUS THERMOPHILUS | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1WAM
 
 | |
1WAN
 
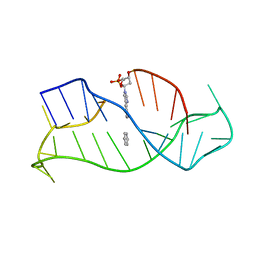 | | DNA DTA TRIPLEX, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*AP*GP*AP*AP*CP*CP*CP*CP*TP*TP*CP*TP*AP*TP*CP*TP*TP*AP*TP*AP*TP*CP*TP*(D3)P*TP*CP*TP*T)-3') | | Authors: | Wang, E, Koshlap, K.M, Gillespie, P, Dervan, P.B, Feigon, J. | | Deposit date: | 1996-01-14 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pyrimidine-purine-pyrimidine triplex containing the sequence-specific intercalating non-natural base D3.
J.Mol.Biol., 257, 1996
|
|
1WAO
 
 | | PP5 structure | | Descriptor: | MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Barford, D. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Tpr Domain-Mediated Regulation of Protein Phosphatase 5
Embo J., 24, 2005
|
|
1WAP
 
 | |
1WAQ
 
 | |
1WAR
 
 | | Recombinant Human Purple Acid Phosphatase expressed in Pichia Pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, HUMAN PURPLE ACID PHOSPHATASE, ... | | Authors: | Duff, A.P, Langley, D.B, Han, R, Averill, B.A, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-10-28 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structures of Recombinant Human Purple Acid Phosphatase with and without an Inhibitory Conformation of the Repression Loop.
J.Mol.Biol., 351, 2005
|
|
1WAS
 
 | |
1WAT
 
 | |
