5H11
 
 | |
5E2B
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
7CFU
 
 | |
2B38
 
 | | Solution structure of kalata B8 | | Descriptor: | kalata B8 | | Authors: | Daly, N.L, Clark, R.J, Plan, M.R, Craik, D.J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif
Biochem.J., 393, 2006
|
|
8Y2O
 
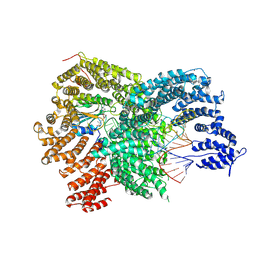 | | The Cryo-EM structure of human tRNA methyltransferase FTSJ1-THADA with substrate tRNA and S-adenosyl homocysteine (SAH) | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Ishiguro, K, Fujimura, A, Shirouzu, M. | | Deposit date: | 2024-01-26 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into tRNA recognition of the human FTSJ1-THADA complex.
Commun Biol, 8, 2025
|
|
4P2L
 
 | | Quiescin Sulfhydryl Oxidase from Rattus norvegicus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfhydryl oxidase 1 | | Authors: | Gat, Y, Fass, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzyme structure captures four cysteines aligned for disulfide relay.
Protein Sci., 23, 2014
|
|
5HZM
 
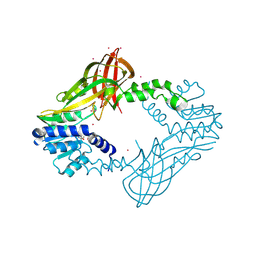 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
5KUP
 
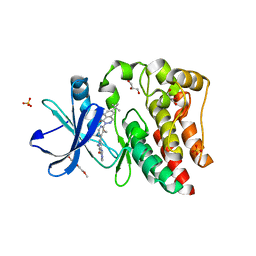 | | Bruton's tyrosine kinase (BTK) with pyridazinone compound 9 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 6-~{tert}-butyl-8-fluoranyl-2-[3-(hydroxymethyl)-4-[1-methyl-6-oxidanylidene-5-(pyrimidin-4-ylamino)pyridin-3-yl]pyridin-2-yl]phthalazin-1-one, GLYCEROL, ... | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Battling Btk Mutants With Noncovalent Inhibitors That Overcome Cys481 and Thr474 Mutations.
Acs Chem.Biol., 11, 2016
|
|
1QOA
 
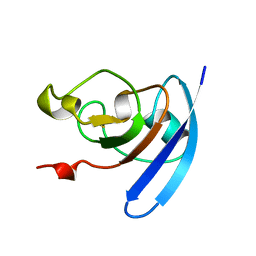 | | FERREDOXIN MUTATION C49S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron-sulfur cluster cysteine-to-serine mutants of Anabaena -2Fe-2S- ferredoxin exhibit unexpected redox properties and are competent in electron transfer to ferredoxin:NADP+ reductase.
Biochemistry, 36, 1997
|
|
5W32
 
 | |
5W3C
 
 | |
3G2P
 
 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-homocysteine (SAH) | | Descriptor: | PCZA361.24, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
4RG1
 
 | | Methyltransferase domain of C9orf114 | | Descriptor: | C9orf114, POLYETHYLENE GLYCOL (N=34), S-1,2-PROPANEDIOL, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of Human C9orf114 in complex with S-adenosyl-homocysteine
To be Published
|
|
1X19
 
 | | Crystal structure of BchU involved in bacteriochlorophyll c biosynthesis | | Descriptor: | CrtF-related protein, SULFATE ION | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-02 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
5W2V
 
 | |
1X1A
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-methionine | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1R8G
 
 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
3VAX
 
 | | Crystal structure of DndA from streptomyces lividans | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein dndA | | Authors: | Zhang, Z, Chen, F, Lin, K, Wu, G. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation
Plos One, 7, 2012
|
|
7MJ1
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with NAD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ0
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with adenosine monophosphate AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ2
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zn | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4G41
 
 | | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Ponniah, K, Norris, G.E, Anderson, B.F, Brown, R.L, Tyler, P.C, Evans, G.B, Frohlich, R. | | Deposit date: | 2012-07-15 | | Release date: | 2012-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin;
TO BE PUBLISHED
|
|
10GS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH TER117 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, L-gamma-glutamyl-S-benzyl-N-[(S)-carboxy(phenyl)methyl]-L-cysteinamide | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
9GIO
 
 | |
3VC2
 
 | | Crystal structure of geranyl diphosphate C-methyltransferase from Streptomyces coelicolor A3(2) in complex with Mg2+, geranyl diphosphate, and S-adenosyl-L-homocysteine | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2012-01-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure of Geranyl Diphosphate C-Methyltransferase from Streptomyces coelicolor and Implications for the Mechanism of Isoprenoid Modification.
Biochemistry, 51, 2012
|
|
