6M2Z
 
 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
6PJL
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-95 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-alloisoleucyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8DT8
 
 | | LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LM18 nanobody, Nb136 nanobody, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PK0
 
 | | Crystal Structure of OXA-48 with Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
6UPY
 
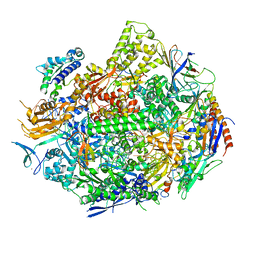 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 2E | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MM8
 
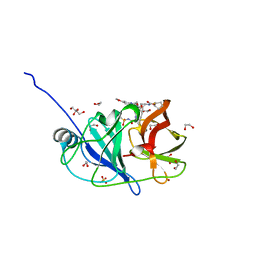 | | Crystal structure of HCV NS3/4A protease in complex with NR02-08 | | Descriptor: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6M0Q
 
 | | Hydroxylamine oxidoreductase from Nitrosomonas europaea | | Descriptor: | Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
8E8Y
 
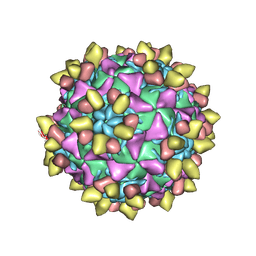 | | 9H2 Fab-Sabin poliovirus 2 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
6M0P
 
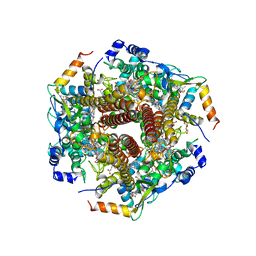 | | Hydroxylamine oxidoreductase in complex with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
6ZRG
 
 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed doripenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-({[amino(dihydroxy)methyl]amino}methyl)pyrrolidin-3-yl]sulfanyl}-5-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6UX2
 
 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7MM2
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-61 | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4a protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
8EB9
 
 | |
6PKH
 
 | |
6ZR6
 
 | |
7MMF
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-60 | | Descriptor: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6M2Y
 
 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
8E1D
 
 | |
6PKQ
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.85A resolution (#11) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6UXK
 
 | |
8DMP
 
 | |
6ZRH
 
 | | Crystal structure of OXA-10loop24 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6UXW
 
 | | SWI/SNF nucleosome complex with ADP-BeFx | | Descriptor: | 601 sequence bottom strand, 601 sequence top strand, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Han, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.96 Å) | | Cite: | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
7MMJ
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-08 | | Descriptor: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
6PL1
 
 | | TRK-A IN COMPLEX WITH LIGAND 1B | | Descriptor: | High affinity nerve growth factor receptor, N-(5-{[(7-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-2-yl)methyl]sulfanyl}-1,3,4-thiadiazol-2-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Subramanian, G. | | Deposit date: | 2019-06-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification and characterization of hTrkA type 2 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
