4JR7
 
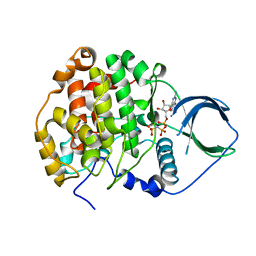 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | Descriptor: | Casein kinase II subunit alpha, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2J78
 
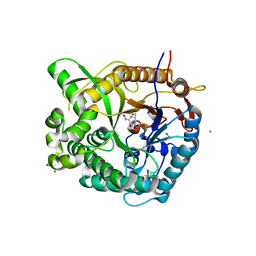 | | Beta-glucosidase from Thermotoga maritima in complex with gluco- hydroximolactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
2J7D
 
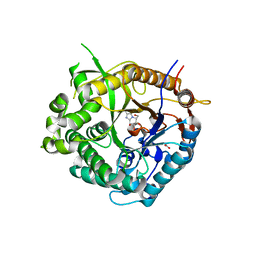 | | Beta-glucosidase from Thermotoga maritima in complex with methoxycarbonyl-substituted glucoimidazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
2IYR
 
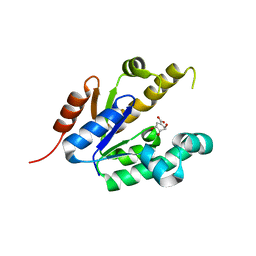 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYW
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with MgATP, open LID (conf. B) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
6XX9
 
 | | Arabidopsis thaliana Casein Kinase 2 (CK2) alpha-1 crystal form III | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Casein kinase II subunit alpha-1, SULFATE ION | | Authors: | Demulder, M, De Veylder, L, Loris, R. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of Arabidopsis thaliana casein kinase 2 alpha 1.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2IYQ
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate and ADP | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IXZ
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*CP*UP*GP*UP*GP*CP*CP)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2JH5
 
 | | Human Thrombin Hirugen Inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Senger, S, Chan, C, Convery, M.A, Hubbard, J.A, Young, R.J, Shah, G.P, Watson, N.S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JH0
 
 | | Human Thrombin Hirugen Inhibitor complex | | Descriptor: | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Senger, S, Chan, C, Convery, M.A, Hubbard, J.A, Young, R.J, Shah, G.P, Watson, N.S. | | Deposit date: | 2007-02-19 | | Release date: | 2007-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JU7
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Protein Only | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
2JCS
 
 | | Active site mutant of dNK from D. melanogaster with dTTP bound | | Descriptor: | DEOXYNUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Egeblad-Welin, L, Sonntag, Y, Eklund, H, Munch-Petersen, B. | | Deposit date: | 2007-01-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Studies of Active-Site Mutants from Drosophila Melanogaster Deoxyribonucleoside Kinase. Investigations of the Putative Catalytic Glutamate- Arginine Pair and of Residues Responsible for Substrate Specificity.
FEBS J., 274, 2007
|
|
2JBK
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with quisqualic acid (quisqualate, alpha-amino-3,5-dioxo-1,2,4- oxadiazolidine-2-propanoic acid) | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2MLH
 
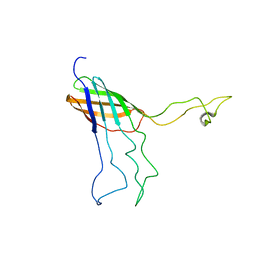 | | NMR Solution Structure of Opa60 from N. Gonorrhoeae in FC-12 Micelles | | Descriptor: | Opacity protein opA60 | | Authors: | Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M, Columbus, L. | | Deposit date: | 2014-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the neisserial outer membrane protein opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2LWA
 
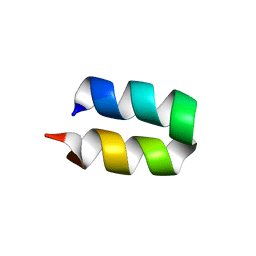 | | Conformational ensemble for the G8A mutant of the influenza hemagglutinin fusion peptide | | Descriptor: | HEMAGGLUTININ FUSION PEPTIDE G8A MUTANT | | Authors: | Lorieau, J.L, Louis, J.M, Schwieters, C.D, Bax, A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2M4Z
 
 | |
6FG5
 
 | |
2M4M
 
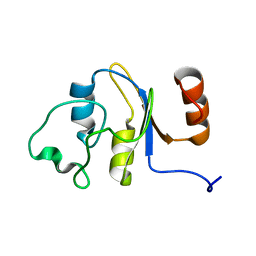 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
8VU6
 
 | |
8VU8
 
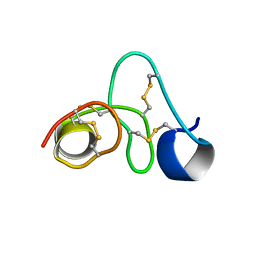 | |
8VU7
 
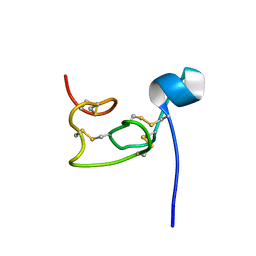 | |
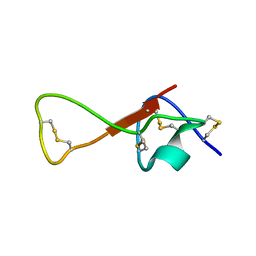
2M36
 
 | |
2MES
 
 | |
2M8P
 
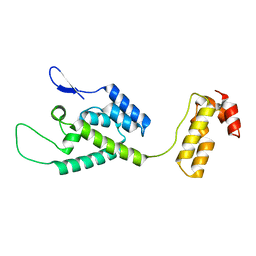 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M4N
 
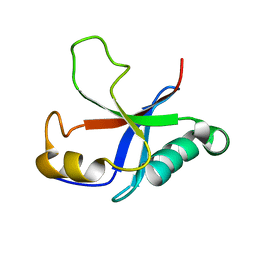 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
