3LGV
 
 | | H198P mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
2FP4
 
 | |
5T75
 
 | | Human carbonic anhydrase II G132C_C206S double mutant in complex with SA-2 | | Descriptor: | 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
2IVU
 
 | |
1RX2
 
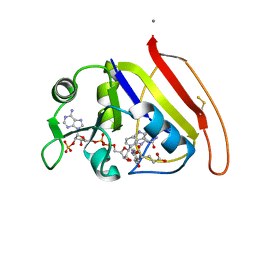 | |
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
7SE6
 
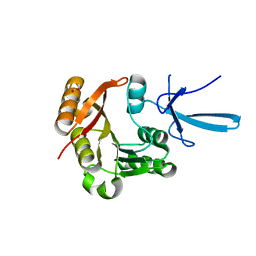 | | Crystal structure of human Fibrillarin in ligand-free state | | Descriptor: | FORMIC ACID, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
8E9N
 
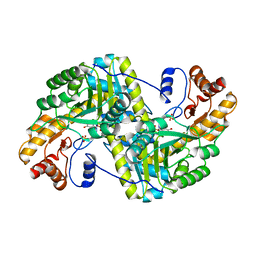 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9T
 
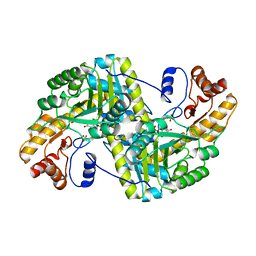 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9L
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9R
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9V
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E4L
 
 | | The open state mouse TRPM8 structure in complex with the cooling agonist C3, AITC, and PI(4,5)P2 | | Descriptor: | 3-isothiocyanatoprop-1-ene, CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
7TXP
 
 | | X-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in complex with TDP-4-amino-4,6-dideoxy-D-glucose | | Descriptor: | SODIUM ION, VioB, dTDP-4-amino-4,6-dideoxyglucose | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
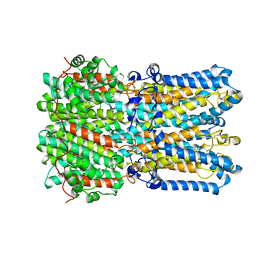
6N25
 
 | |
4FDO
 
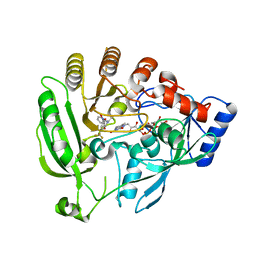 | | Mycobacterium tuberculosis DprE1 in complex with CT319 | | Descriptor: | 3-nitro-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, oxidoreductase DprE1 | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4PE5
 
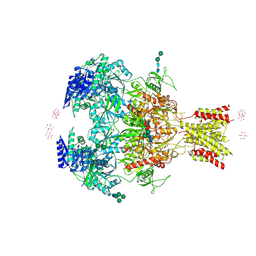 | | Crystal Structure of GluN1a/GluN2B NMDA Receptor Ion Channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, ... | | Authors: | Karakas, E, Furukawa, H. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystal structure of a heterotetrameric NMDA receptor ion channel.
Science, 344, 2014
|
|
3G5Z
 
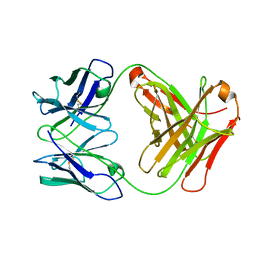 | |
3F5E
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 2'F-3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
4RR9
 
 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Ser3AA (snapshot 4) | | Descriptor: | MAGNESIUM ION, Probable threonine--tRNA ligase 2, SERINE-3'-AMINOADENOSINE | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRL
 
 | |
2JJ3
 
 | | Estrogen receptor beta ligand binding domain in complex with a Benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, ESTROGEN RECEPTOR BETA | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Benzopyrans as Selective Estrogen Receptor Beta Agonists (Serbas). Part 4: Functionalization of the Benzopyran A-Ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3F5A
 
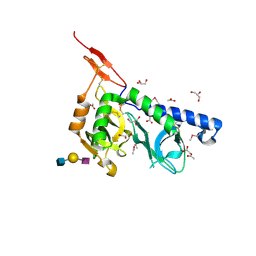 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
4RRD
 
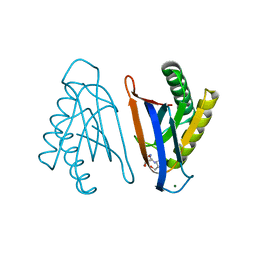 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Thr3AA (snapshot 4) | | Descriptor: | 3'-deoxy-3'-(L-threonylamino)adenosine, MAGNESIUM ION, Probable threonine--tRNA ligase 2 | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
5IPS
 
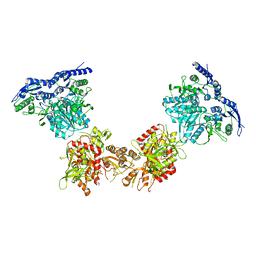 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 4 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
