4UDT
 
 | | T cell receptor (TRAV22,TRBV7-9) structure | | Descriptor: | GLYCEROL, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
5QIE
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z2856434894 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1, ~{N},~{N}-dimethyl-4-[(propan-2-ylamino)methyl]aniline | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QIF
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QIC
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z30620520 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1, cyclopropyl-[4-(4-fluorophenyl)piperazin-1-yl]methanone | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QIH
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z2697514548 | | Descriptor: | 1-methylindazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QID
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z1787627869 | | Descriptor: | 5-chloranyl-~{N}-methyl-~{N}-[[(3~{S})-oxolan-3-yl]methyl]pyrimidin-4-amine, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
6P9U
 
 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
8BEO
 
 | | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, ... | | Authors: | Shaanan, B, Binshtein, E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP
Not Published
|
|
6GF3
 
 | | Tubulin-Jerantinine B acetate complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Smedley, C.J, Stanley, P.A, Qazzaz, M.E, Prota, A.E, Olieric, N, Collins, H, Eastman, H, Barrow, A.S, Lim, K.-H, Kam, T.-S, Smith, B.J, Duivenvoorden, H.M, Parker, B.S, Bradshaw, T.D, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Syntheses of (-)-Jerantinines A & E and Structural Characterisation of the Jerantinine-Tubulin Complex at the Colchicine Binding Site.
Sci Rep, 8, 2018
|
|
6PX5
 
 | | CRYSTAL STRUCTURE OF HUMAN MEIZOTHROMBIN DESF1 MUTANT S195A bound with PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Gistover, N, Di Cera, E. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
8G02
 
 | | YES Complex - E. coli MraY, Protein E PhiX174, E. coli SlyD | | Descriptor: | Lysis protein E, Peptidyl-prolyl cis-trans isomerase, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Li, Y.E. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
8CAG
 
 | | Hypoxanthine-guanine phosphoribosyltransferase from E. coli | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Kostromina, M.A, Zayats, E.A, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hypoxanthine-guanine phosphoribosyltransferase from E. coli
To Be Published
|
|
6G9P
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6OUL
 
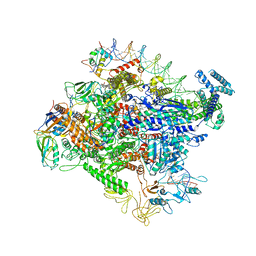 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound to rpsTP2 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
3DNF
 
 | |
6G9S
 
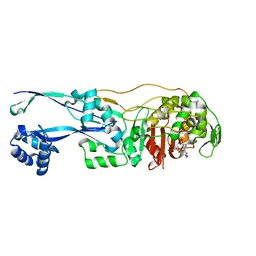 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G9F
 
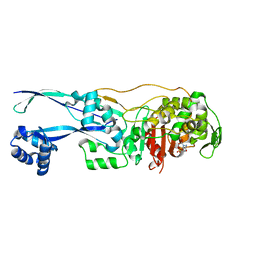 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
8C8F
 
 | |
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.14885437 Å) | | Cite: | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
7Q9N
 
 | | Transthyretin complexed with (E)-4-(2-(naphthalen-2-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-2-ylethenyl]benzene-1,2-diol, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7Q9L
 
 | | Transthyretin complexed with (E)-4-(2-(naphthalen-1-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-1-ylethenyl]benzene-1,2-diol, GLYCEROL, SODIUM ION, ... | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7QFM
 
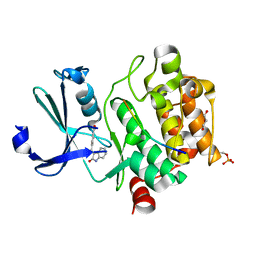 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
4Y4N
 
 | | Thiazole synthase Thi4 from Methanococcus igneus | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (II) ION, Putative ribose 1,5-bisphosphate isomerase | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
6NPF
 
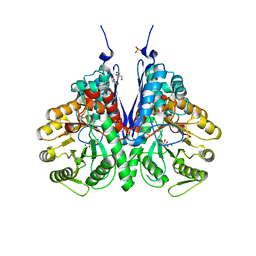 | | Structure of E.coli enolase in complex with an analog of the natural product SF-2312 metabolite. | | Descriptor: | Enolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Erlandsen, H, Krucinska, J, Lombardo, M, Wright, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6OH6
 
 | |
