7N72
 
 | |
7N78
 
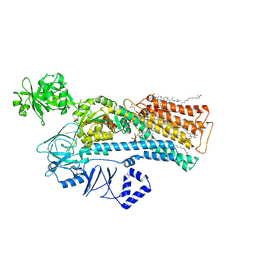 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sim, S.I, Park, E. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of polyamine transport by human ATP13A2 (PARK9).
Mol.Cell, 81, 2021
|
|
7N74
 
 | |
7N76
 
 | |
7N75
 
 | |
7N73
 
 | |
7BSS
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in E1AlF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, CDC50A, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BH2
 
 | | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BSQ
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in E1AlF-ADP state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BSU
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-bound E2BeF state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BH1
 
 | | Cryo-EM Structure of KdpFABC in E1 state with K | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BSP
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in E1-AMPPCP state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BT2
 
 | | Crystal structure of the SERCA2a in the E2.ATP state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kabashima, Y, Ogawa, H, Nakajima, R, Toyoshima, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00002861 Å) | | Cite: | What ATP binding does to the Ca2+pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BGY
 
 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BSV
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-occluded E2-AlF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, CDC50A, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BSW
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdEtn-occluded E2-AlF state | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
6HXB
 
 | | SERCA2a from pig heart | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Sitsel, A, Andersen, J.L, Nissen, P, Olesen, C. | | Deposit date: | 2018-10-16 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of the heart specific SERCA2a Ca 2+ -ATPase.
Embo J., 38, 2019
|
|
6JJU
 
 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | Deposit date: | 2019-02-27 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
6K7H
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7N
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7I
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6JXI
 
 | | Rb+-bound E2-MgF state of the gastric proton pump (Tyr799Trp) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Irie, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single K + -binding site in the crystal structure of the gastric proton pump.
Elife, 8, 2019
|
|
6K7J
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6HEF
 
 | | Room temperature structure of the (SR)Ca2+-ATPase Ca2-E1-CaAMPPCP form | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Hjorth-Jensen, S, Sorensen, T.L.M, Oksanen, E, Andersen, J.L, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2018-08-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | Membrane-protein crystals for neutron diffraction.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6HRA
 
 | | Cryo-EM structure of the KdpFABC complex in an E1 outward-facing state (state 1) | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Stock, C, Hielkema, L, Tascon, I, Wunnicke, D, Oostergetel, G.T, Azkargorta, M, Paulino, C, Haenelt, I. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Nat Commun, 9, 2018
|
|
