2QS2
 
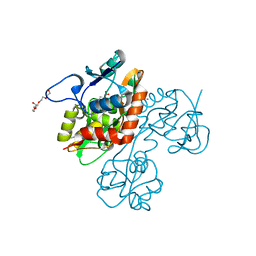 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2RCA
 
 | |
2QS3
 
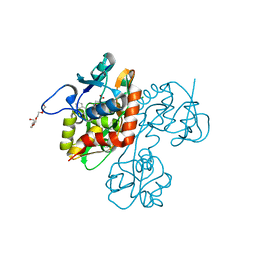 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP316 at 1.76 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-5-phenylthiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function.
Neuropharmacology, 56, 2009
|
|
2RCB
 
 | |
2RC7
 
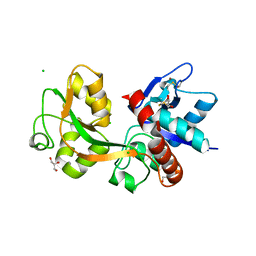 | |
4ISU
 
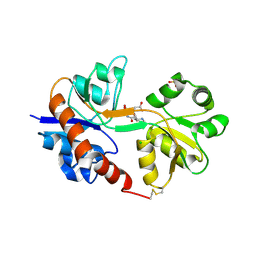 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | Descriptor: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
4JWX
 
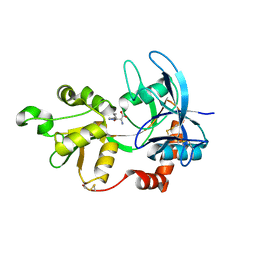 | | GluN2A ligand-binding core in complex with propyl-NHP5G | | Descriptor: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, GluN2A | | Authors: | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
4KCC
 
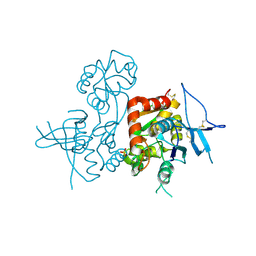 | | Crystal Structure of the NMDA Receptor GluN1 Ligand Binding Domain Apo State | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, PHOSPHATE ION | | Authors: | Berger, A.J, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
4L17
 
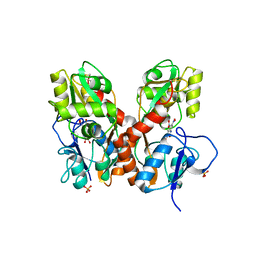 | | GluA2-L483Y-A665C ligand-binding domain in complex with the antagonist DNQX | | Descriptor: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | Authors: | Lau, A.Y, Blachowicz, L, Roux, B. | | Deposit date: | 2013-06-02 | | Release date: | 2013-08-14 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A conformational intermediate in glutamate receptor activation.
Neuron, 79, 2013
|
|
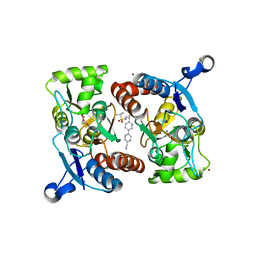
4LZ5
 
 | |
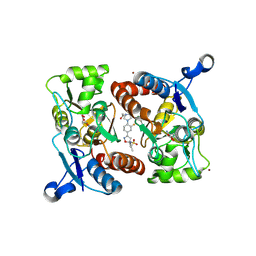
4LZ8
 
 | |
4MF3
 
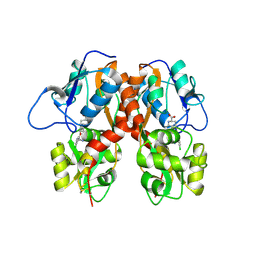 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LZ7
 
 | |
4MH5
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
4KCD
 
 | | Crystal Structure of the NMDA Receptor GluN3A Ligand Binding Domain Apo State | | Descriptor: | GLYCEROL, Glutamate receptor ionotropic, NMDA 3A | | Authors: | Yao, Y, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
7EOQ
 
 | |
7EOT
 
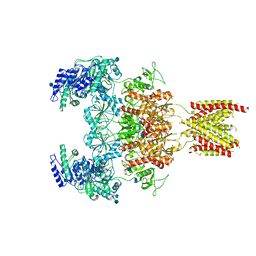 | |
7EOU
 
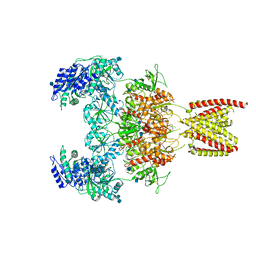 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EOS
 
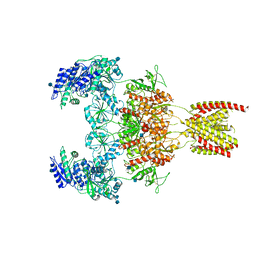 | |
7EOR
 
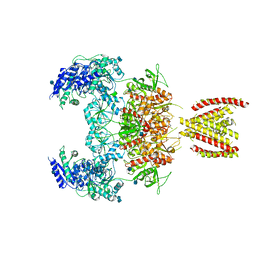 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EU8
 
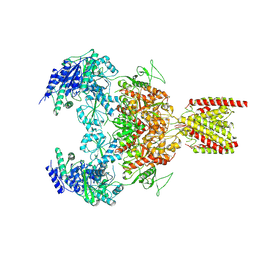 | | Structure of the human GluN1-GluN2B NMDA receptor in complex with S-ketamine,glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, T, Zhang, Y, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
7F3O
 
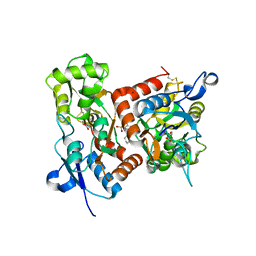 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-653 | | Descriptor: | 7-(4-cyclohexyloxyphenyl)-9-methyl-4$l^{6}-thia-1$l^{4},5,8-triazabicyclo[4.4.0]deca-1(10),6,8-triene 4,4-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2021-06-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strictly regulated agonist-dependent activation of AMPA-R is the key characteristic of TAK-653 for robust synaptic responses and cognitive improvement.
Sci Rep, 11, 2021
|
|
7EU7
 
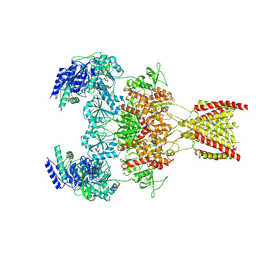 | | Structure of the human GluN1-GluN2A NMDA receptor in complex with S-ketamine, glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, Y, Zhang, T, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-04 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
4KFQ
 
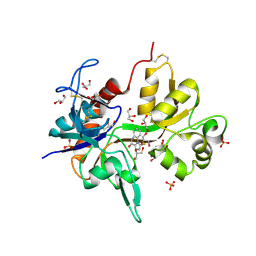 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | Descriptor: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
4JWY
 
 | | GluN2D ligand-binding core in complex with propyl-NHP5G | | Descriptor: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, Glutamate receptor ionotropic, NMDA 2D | | Authors: | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
