6D9J
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-sitetRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
2H5E
 
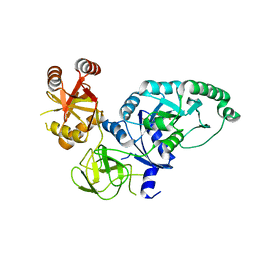 | | Crystal structure of E.coli polypeptide release factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor RF-3 | | Authors: | Song, H.W, Zhou, Z.H. | | Deposit date: | 2006-05-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
2HDN
 
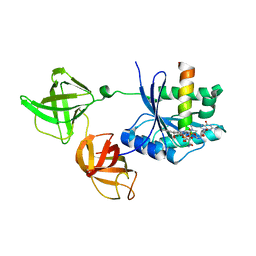 | | Trypsin-modified Elongation Factor Tu in complex with tetracycline at 2.8 Angstrom resolution | | Descriptor: | Elongation factor EF-Tu, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mui, S, Heffron, S.E, Aorora, A, Abel, K, Bergmann, E, Jurnak, F. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular complementarity between tetracycline and the GTPase active site of elongation factor Tu.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HCJ
 
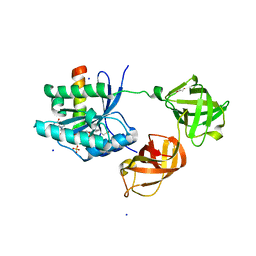 | | Trypsin-modified Elongation Factor Tu in complex with tetracycline | | Descriptor: | GLYOXYLIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mui, S, Heffron, S.E, Aorora, A, Abel, K, Bergmann, E, Jurnak, F. | | Deposit date: | 2006-06-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular complementarity between tetracycline and the GTPase active site of elongation factor Tu.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8HL2
 
 | |
1FNM
 
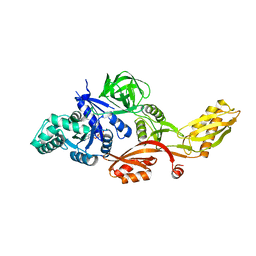 | | STRUCTURE OF THERMUS THERMOPHILUS EF-G H573A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Laurberg, M, Kristensen, O, Martemyanov, K, Gudkov, A.T, Nagaev, I, Hughes, D, Liljas, A. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site.
J.Mol.Biol., 303, 2000
|
|
8HL4
 
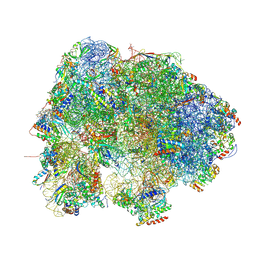 | |
6RW5
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF2 and IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
8H6E
 
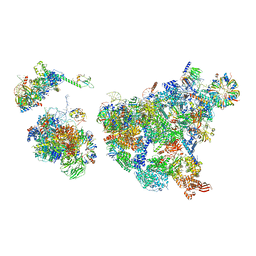 | | Cryo-EM structure of human exon-defined spliceosome in the late pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6L
 
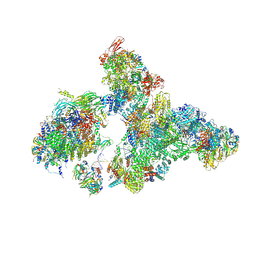 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6K
 
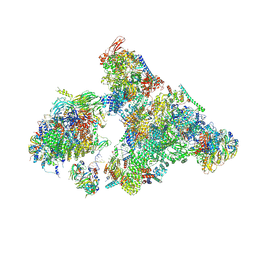 | | Cryo-EM structure of human exon-defined spliceosome in the mature B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6J
 
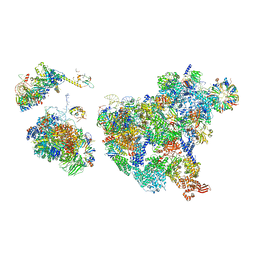 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
1G7S
 
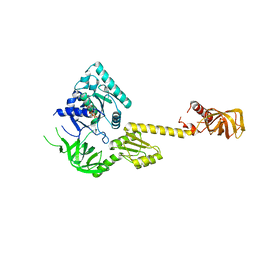 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G7R
 
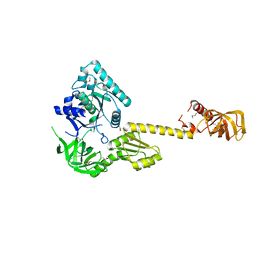 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B | | Descriptor: | TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
8DMF
 
 | | Cryo-EM structure of the ribosome-bound Bacteroides thetaiotaomicron EF-G2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tetracycline resistance protein TetQ | | Authors: | Wang, C, Han, W, Groisman, E.A, Liu, J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.
Embo J., 2022
|
|
1G7C
 
 | | YEAST EEF1A:EEF1BA IN COMPLEX WITH GDPNP | | Descriptor: | ELONGATION FACTOR 1-ALPHA, ELONGATION FACTOR 1-BETA, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
1G7T
 
 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HA3
 
 | | ELONGATION FACTOR TU IN COMPLEX WITH aurodox | | Descriptor: | BETA-MERCAPTOETHANOL, ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vogeley, L, Palm, G.J, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Change of Elongation Factor TU Induced by Antibiotic Binding: Crystal Structure of the Complex between EF-TU:Gdp and Aurodox
J.Biol.Chem., 276, 2001
|
|
1N0U
 
 | | Crystal structure of yeast elongation factor 2 in complex with sordarin | | Descriptor: | Elongation factor 2, [1R-(1.ALPHA.,3A.BETA.,4.BETA.,4A.BETA.,7.BETA.,7A.ALPHA.,8A.BETA.)]8A-[(6-DEOXY-4-O-METHYL-BETA-D-ALTROPYRANOSYLOXY)METHYL]-4-FORMYL-4,4A,5,6,7,7A,8,8A-OCTAHYDRO-7-METHYL-3-(1-METHYLETHYL)-1,4-METHANO-S-INDACENE-3A(1H)-CARBOXYLIC ACID | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat. Struct. Biol., 10, 2003
|
|
6SW9
 
 | | IC2A model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
8CEH
 
 | | Translocation intermediate 4 (TI-4) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CKU
 
 | | Translocation intermediate 1 (TI-1*) of 80S S. cerevisiae ribosome with ligands and eEF2 in the absence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CIV
 
 | | Translocation intermediate 5 (TI-5) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
6U45
 
 | |
8EWH
 
 | | Salmonella typhimurium GTPase BIPA | | Descriptor: | 50S ribosomal subunit assembly factor BipA, SODIUM ION | | Authors: | Brown, R.S, deLivron, M.A, Robinson, V.L. | | Deposit date: | 2022-10-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic and Biochemical Characterization of the GTPase and Ribosome Binding Properties of Salmonella typhimuirum BipA
J Biomol Struct Dyn., 24:6, 2007
|
|
