4N4X
 
 | | Crystal Structure of the MBP fused human SPLUNC1 (native form) | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Maltose-binding periplasmic/Palate lung and nasal epithelium clone fusion protein | | Authors: | Ning, F, Wang, C, Niu, L, Chu, H.W, Zhang, G. | | Deposit date: | 2013-10-08 | | Release date: | 2014-09-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Lipid ligands of human SPLUNC1
To be Published
|
|
5ZNZ
 
 | | Structure of mDR3 DD with MBP tag mutant-I387V | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Jin, T, Yin, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
7MHW
 
 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
6D67
 
 | | Crystal structure of the human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion (maltose bound form) in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Designed AR protein mbp3_16, ... | | Authors: | Gumpena, R, Lountos, G.T, Waugh, D.S. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | Authors: | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
4GIZ
 
 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
5AZ6
 
 | |
4KYE
 
 | | Partial Structure of the C-terminal domain of the HPIV4B phosphoprotein, fused to MBP. | | Descriptor: | Maltose-binding periplasmic protein, Phosphoprotein, chimeric construct, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
8C5L
 
 | |
1PEB
 
 | | LIGAND-FREE HIGH-AFFINITY MALTOSE-BINDING PROTEIN | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
4XAI
 
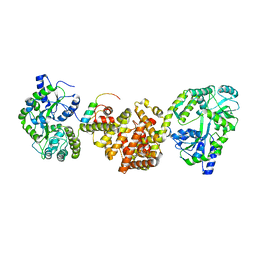 | | Crystal Structure of red flour beetle NR2E1/TLX | | Descriptor: | Grunge, isoform J, Maltose-binding periplasmic protein,Tailless ortholog, ... | | Authors: | Zhi, X, Zhou, E, Xu, E. | | Deposit date: | 2014-12-14 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
3MP1
 
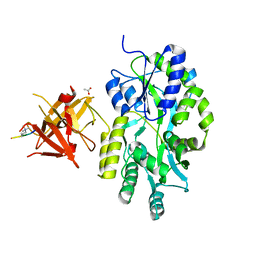 | | Complex structure of Sgf29 and trimethylated H3K4 | | Descriptor: | ACETATE ION, H3K4me3 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, ... | | Authors: | Li, J, Ruan, J, Wu, M, Xue, X, Zang, J. | | Deposit date: | 2010-04-24 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
7U0G
 
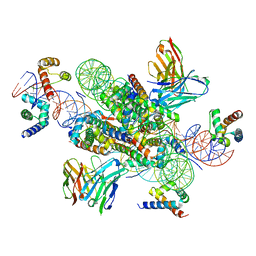 | | structure of LIN28b nucleosome bound 3 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
7U0I
 
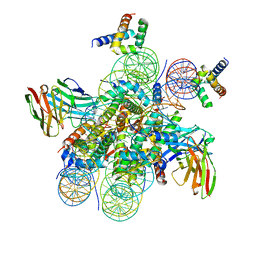 | | Structure of LIN28b nucleosome bound 2 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Tengfei, L, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
8J25
 
 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
5BK2
 
 | |
1N3W
 
 | | Engineered High-Affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2002-10-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
3F5F
 
 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8TLV
 
 | |
5YGS
 
 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
4O2X
 
 | | Structure of a malarial protein | | Descriptor: | Maltose-binding periplasmic protein, ATP-dependent Clp protease adaptor protein ClpS containing protein chimeric construct | | Authors: | AhYoung, A.P, Koehl, A, Cascio, D, Egea, P.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a putative ClpS N-end rule adaptor protein from the malaria pathogen Plasmodium falciparum.
Protein Sci., 25, 2016
|
|
6DM8
 
 | | Understanding the Species Selectivity of Myeloid cell leukemia-1 (Mcl-1) inhibitors | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog - MBP chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding the Species Selectivity of Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors.
Biochemistry, 57, 2018
|
|
6QUG
 
 | | GHK tagged MBP-Nup98(1-29) | | Descriptor: | COPPER (II) ION, Maltodextrin-binding protein,Nucleoporin, putative, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5HZV
 
 | | Crystal structure of the zona pellucida module of human endoglin/CD105 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Endoglin, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
4LOG
 
 | | The crystal structure of the orphan nuclear receptor PNR ligand binding domain fused with MBP | | Descriptor: | Maltose ABC transporter periplasmic protein and NR2E3 protein chimeric construct | | Authors: | Tan, M.E, Zhou, X.E, Soon, F.-F, Li, X, Li, J, Yong, E.-L, Melcher, K, Xu, H.E. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Orphan Nuclear Receptor NR2E3/PNR Ligand Binding Domain Reveals a Dimeric Auto-Repressed Conformation.
Plos One, 8, 2013
|
|
