6CXS
 
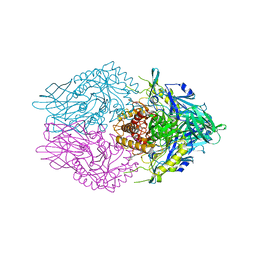 | | Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine | | Descriptor: | 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine, Beta-glucuronidase, Maltose/maltodextrin-binding periplasmic protein | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted inhibition of gut bacterial beta-glucuronidase activity enhances anticancer drug efficacy.
Proc.Natl.Acad.Sci.USA, 2020
|
|
7XQC
 
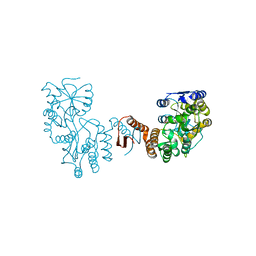 | |
4BL9
 
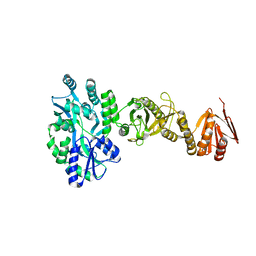 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLB
 
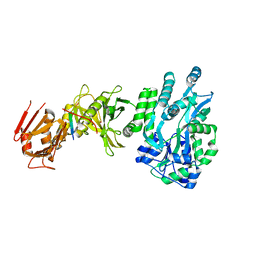 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2R6G
 
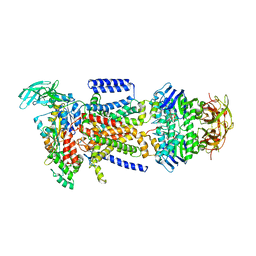 | | The Crystal Structure of the E. coli Maltose Transporter | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Maltose transport system permease protein malF, Maltose transport system permease protein malG, ... | | Authors: | Oldham, M.L, Khare, D, Quiocho, F.A, Davidson, A.L, Chen, J. | | Deposit date: | 2007-09-05 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a catalytic intermediate of the maltose transporter.
Nature, 450, 2007
|
|
4BLD
 
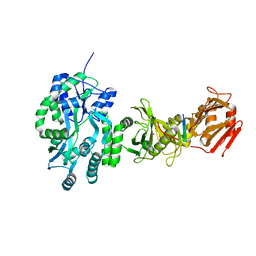 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6KXG
 
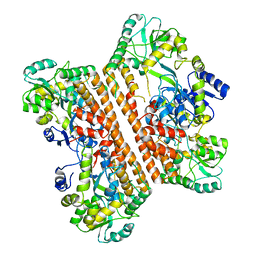 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
4FED
 
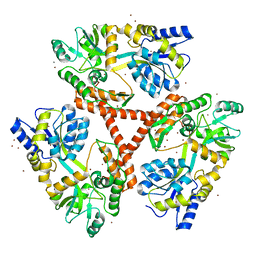 | | Crystal Structure of Htt36Q3H | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
8DEI
 
 | | Structure of the Cac1 KER domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Maltodextrin-binding protein,Chromatin assembly factor 1 subunit p90 fusion, ... | | Authors: | Rosas, R, Churchill, M.E.A. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A novel single alpha-helix DNA-binding domain in CAF-1 promotes gene silencing and DNA damage survival through tetrasome-length DNA selectivity and spacer function.
Elife, 12, 2023
|
|
6EG3
 
 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6DD5
 
 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
5DIS
 
 | | Crystal structure of a CRM1-RanGTP-SPN1 export complex bound to a 113 amino acid FG-repeat containing fragment of Nup214 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Monecke, T, Port, S.A, Dickmanns, A, Kehlenbach, R.H, Ficner, R. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export.
Cell Rep, 13, 2015
|
|
7FBD
 
 | | De novo design protein D53 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D53 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
4QSZ
 
 | |
7BG0
 
 | | Fusion of MBP and the backbone of the long-acting amylin analog AM833. | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Islet amyloid polypeptide, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Johansson, E. | | Deposit date: | 2021-01-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Development of Cagrilintide, a Long-Acting Amylin Analogue.
J.Med.Chem., 64, 2021
|
|
4KHZ
 
 | | Crystal structure of the maltose-binding protein/maltose transporter complex in an pre-translocation conformation bound to maltoheptaose | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Binding-protein-dependent transport systems inner membrane component, Maltose transport system permease protein MalF, ... | | Authors: | Oldham, M.L, Chen, S, Chen, J. | | Deposit date: | 2013-05-01 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for substrate specificity in the Escherichia coli maltose transport system.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3D4C
 
 | |
1A7L
 
 | | DOMINANT B-CELL EPITOPE FROM THE PRES2 REGION OF HEPATITIS B VIRUS IN THE FORM OF AN INSERTED PEPTIDE SEGMENT IN MALTODEXTRIN-BINDING PROTEIN | | Descriptor: | MALE-B363, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Vulliez-Lenormand, B, Lema, F, Bentley, G.A. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a dominant B-cell epitope from the preS2 region of hepatitis B virus in the form of an inserted peptide segment in maltodextrin-binding protein.
J.Mol.Biol., 280, 1998
|
|
5IIC
 
 | | Crystal structure of red abalone VERL repeat 3 at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6HDB
 
 | | Crystal structure of the potassium channel MtTMEM175 with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Rang, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
3PUZ
 
 | |
5W0U
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
6I4Y
 
 | | X-ray structure of the human mitochondrial PRELID3b-TRIAP1 complex | | Descriptor: | Maltose transport system, substrate-binding protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3B, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
